The conserved Fanconi anemia nuclease Fan1 and the SUMO E3 ligase Pli1 act in two novel Pso2-independent pathways of DNA interstrand crosslink repair in yeast
- PMID: 24192486
- PMCID: PMC4045212
- DOI: 10.1016/j.dnarep.2013.10.003
The conserved Fanconi anemia nuclease Fan1 and the SUMO E3 ligase Pli1 act in two novel Pso2-independent pathways of DNA interstrand crosslink repair in yeast
Erratum in
-
Corrigendum to "The conserved Fanconi anemia nuclease Fan1 and the SUMO E3 ligase Pli1 act in two novel Pso2-independent pathways of DNA interstrand crosslink repair in yeast" [DNA Repair 12 (December (12)) (2013) 1011-1023].DNA Repair (Amst). 2014 Dec;24:150. doi: 10.1016/j.dnarep.2014.11.003. Epub 2014 Nov 20. DNA Repair (Amst). 2014. PMID: 28843320 Free PMC article. No abstract available.
Abstract
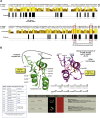
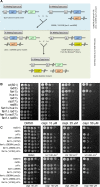
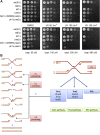
DNA interstrand cross-links (ICLs) represent a physical barrier to the progression of cellular machinery involved in DNA metabolism. Thus, this type of adduct represents a serious threat to genomic stability and as such, several DNA repair pathways have evolved in both higher and lower eukaryotes to identify this type of damage and restore the integrity of the genetic material. Human cells possess a specialized ICL-repair system, the Fanconi anemia (FA) pathway. Conversely yeasts rely on the concerted action of several DNA repair systems. Recent work in higher eukaryotes identified and characterized a novel conserved FA component, FAN1 (Fanconi anemia-associated nuclease 1, or FANCD2/FANCI-associated nuclease 1). In this study, we characterize Fan1 in the yeast Schizosaccharomyces pombe. Using standard genetics, we demonstrate that Fan1 is a key component of a previously unidentified ICL-resolution pathway. Using high-throughput synthetic genetic arrays, we also demonstrate the existence of a third pathway of ICL repair, dependent on the SUMO E3 ligase Pli1. Finally, using sequence-threaded homology models, we predict and validate key residues essential for Fan1 activity in ICL repair.
Keywords: Cisplatin; Epistasis; Genetic screen; ICL; Schizosaccharomyces pombe; Synthetic array.
Copyright © 2013. Published by Elsevier B.V.
Figures



Similar articles
-
Fan1 deficiency results in DNA interstrand cross-link repair defects, enhanced tissue karyomegaly, and organ dysfunction.Genes Dev. 2016 Mar 15;30(6):645-59. doi: 10.1101/gad.276261.115. Genes Dev. 2016. PMID: 26980189 Free PMC article.
-
Corrigendum to "The conserved Fanconi anemia nuclease Fan1 and the SUMO E3 ligase Pli1 act in two novel Pso2-independent pathways of DNA interstrand crosslink repair in yeast" [DNA Repair 12 (December (12)) (2013) 1011-1023].DNA Repair (Amst). 2014 Dec;24:150. doi: 10.1016/j.dnarep.2014.11.003. Epub 2014 Nov 20. DNA Repair (Amst). 2014. PMID: 28843320 Free PMC article. No abstract available.
-
The yeast Hrq1 helicase stimulates Pso2 translesion nuclease activity and thereby promotes DNA interstrand crosslink repair.J Biol Chem. 2020 Jul 3;295(27):8945-8957. doi: 10.1074/jbc.RA120.013626. Epub 2020 May 5. J Biol Chem. 2020. PMID: 32371399 Free PMC article.
-
Ubiquitylation and the Fanconi anemia pathway.FEBS Lett. 2011 Sep 16;585(18):2853-60. doi: 10.1016/j.febslet.2011.04.078. Epub 2011 May 19. FEBS Lett. 2011. PMID: 21605559 Free PMC article. Review.
-
Structural and functional relationships of FAN1.DNA Repair (Amst). 2017 Aug;56:135-143. doi: 10.1016/j.dnarep.2017.06.016. Epub 2017 Jun 9. DNA Repair (Amst). 2017. PMID: 28623094 Review.
Cited by
-
Fan1 deficiency results in DNA interstrand cross-link repair defects, enhanced tissue karyomegaly, and organ dysfunction.Genes Dev. 2016 Mar 15;30(6):645-59. doi: 10.1101/gad.276261.115. Genes Dev. 2016. PMID: 26980189 Free PMC article.
-
Quantitative proteomics and phosphoproteomics profiling of meiotic divisions in the fission yeast Schizosaccharomyces pombe.Sci Rep. 2024 Oct 4;14(1):23105. doi: 10.1038/s41598-024-74523-0. Sci Rep. 2024. PMID: 39367033 Free PMC article.
-
Functions and regulation of the multitasking FANCM family of DNA motor proteins.Genes Dev. 2015 Sep 1;29(17):1777-88. doi: 10.1101/gad.266593.115. Genes Dev. 2015. PMID: 26341555 Free PMC article. Review.
-
Ubiquitylation at the Fork: Making and Breaking Chains to Complete DNA Replication.Int J Mol Sci. 2018 Sep 25;19(10):2909. doi: 10.3390/ijms19102909. Int J Mol Sci. 2018. PMID: 30257459 Free PMC article. Review.
-
FANCD2-Associated Nuclease 1 Partially Compensates for the Lack of Exonuclease 1 in Mismatch Repair.Mol Cell Biol. 2021 Aug 24;41(9):e0030321. doi: 10.1128/MCB.00303-21. Epub 2021 Aug 24. Mol Cell Biol. 2021. PMID: 34228493 Free PMC article.
References
-
- McHugh P.J., Spanswick V.J., Hartley J.A. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. - PubMed
-
- Lehoczký P., McHugh P.J., Chovanec M. DNA interstrand cross-link repair in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2007;31:109–133. - PubMed
-
- Ruhland A., Kircher M., Wilborn F., Brendel M. A yeast mutant specifically sensitive to bifunctional alkylation. Mutat. Res. 1981;91:457–462. - PubMed
-
- Cassier-Chauvat C., Moustacchi E. Allelism between pso1-1 and rev3-1 mutants and between pso2-1 and snm1 mutants in Saccharomyces cerevisiae. Curr. Genet. 1988;13:37–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

