WaveCNV: allele-specific copy number alterations in primary tumors and xenograft models from next-generation sequencing
- PMID: 24192544
- PMCID: PMC3957071
- DOI: 10.1093/bioinformatics/btt611
WaveCNV: allele-specific copy number alterations in primary tumors and xenograft models from next-generation sequencing
Abstract
Motivation: Copy number variations (CNVs) are a major source of genomic variability and are especially significant in cancer. Until recently microarray technologies have been used to characterize CNVs in genomes. However, advances in next-generation sequencing technology offer significant opportunities to deduce copy number directly from genome sequencing data. Unfortunately cancer genomes differ from normal genomes in several aspects that make them far less amenable to copy number detection. For example, cancer genomes are often aneuploid and an admixture of diploid/non-tumor cell fractions. Also patient-derived xenograft models can be laden with mouse contamination that strongly affects accurate assignment of copy number. Hence, there is a need to develop analytical tools that can take into account cancer-specific parameters for detecting CNVs directly from genome sequencing data.
Results: We have developed WaveCNV, a software package to identify copy number alterations by detecting breakpoints of CNVs using translation-invariant discrete wavelet transforms and assign digitized copy numbers to each event using next-generation sequencing data. We also assign alleles specifying the chromosomal ratio following duplication/loss. We verified copy number calls using both microarray (correlation coefficient 0.97) and quantitative polymerase chain reaction (correlation coefficient 0.94) and found them to be highly concordant. We demonstrate its utility in pancreatic primary and xenograft sequencing data.
Availability and implementation: Source code and executables are available at https://github.com/WaveCNV. The segmentation algorithm is implemented in MATLAB, and copy number assignment is implemented Perl.
Contact: lakshmi.muthuswamy@gmail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
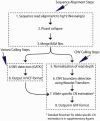

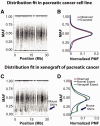


References
-
- Coifman RR, Donoho DL. Translation-invariant de-noising. In: Antoniadis A, Oppenheim G, editors. Wavelets and Statistics. New York: Springer-Verlag; 1995.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

