Genome-wide transcriptional effects of the anti-cancer agent camptothecin
- PMID: 24194914
- PMCID: PMC3806802
- DOI: 10.1371/journal.pone.0078190
Genome-wide transcriptional effects of the anti-cancer agent camptothecin
Abstract
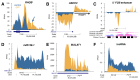
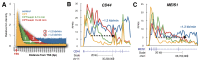
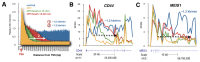
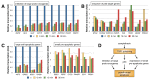
The anti-cancer drug camptothecin inhibits replication and transcription by trapping DNA topoisomerase I (Top1) covalently to DNA in a "cleavable complex". To examine the effects of camptothecin on RNA synthesis genome-wide we used Bru-Seq and show that camptothecin treatment primarily affected transcription elongation. We also observed that camptothecin increased RNA reads past transcription termination sites as well as at enhancer elements. Following removal of camptothecin, transcription spread as a wave from the 5'-end of genes with no recovery of transcription apparent from RNA polymerases stalled in the body of genes. As a result, camptothecin preferentially inhibited the expression of large genes such as proto-oncogenes, and anti-apoptotic genes while smaller ribosomal protein genes, pro-apoptotic genes and p53 target genes showed relative higher expression. Cockayne syndrome group B fibroblasts (CS-B), which are defective in transcription-coupled repair (TCR), showed an RNA synthesis recovery profile similar to normal fibroblasts suggesting that TCR is not involved in the repair of or RNA synthesis recovery from transcription-blocking Top1 lesions. These findings of the effects of camptothecin on transcription have important implications for its anti-cancer activities and may aid in the design of improved combinatorial treatments involving Top1 poisons.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

