Comparative analysis of putative orthologues of mitochondrial import motor subunit: Pam18 and Pam16 in plants
- PMID: 24194927
- PMCID: PMC3806816
- DOI: 10.1371/journal.pone.0078400
Comparative analysis of putative orthologues of mitochondrial import motor subunit: Pam18 and Pam16 in plants
Abstract
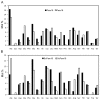
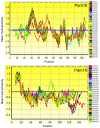
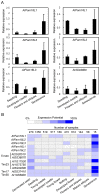
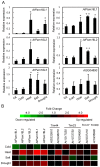
Pam18/Tim14 and Pam16/Tim16, highly conserved proteins among eukaryotes, are two essential subunits of protein import motors localized in the inner mitochondrial membrane. The heterodimer formed by Pam18 and Pam16 via their J-type domains serves a regulatory function in protein translocation. Here, we report that thirty-one Pam18 and twenty-six Pam16 putative orthologues in twelve plant species were identified and analyzed through bioinformatics strategy. Results data revealed that Pam18 and Pam16 were also highly conserved among plants including their J-type domains within the hydrophilic region. Key amino acid residues and an HPD motif of Pam18 were identical among the orthologues except OsPam18L5. N-myristoylation sites of Pam18 and casein kinase II phosphorylation sites of Pam 16 were more abundant, which might be important functional sites. Some Pam18 and Pam16 proteins contained a transmembrane region at the N-terminal region. Sub-cellular prediction results indicated that many orthologues localized at mitochondria. Gene expression analyses revealed that Pam18 and Pam16 in Arabidopsis might play roles in senescence and abiotic stress responses. Our detailed study provides a better understanding of Pam18 and Pam16 in plant kingdom.
Conflict of interest statement
Figures


Similar articles
-
Common functions of the chloroplast and mitochondrial co-chaperones cpDnaJL (CDF1) and mtDnaJ (PAM16) in protein import and ROS scavenging in Arabidopsis thaliana.Commun Integr Biol. 2015 Dec 9;9(5):e1119343. doi: 10.1080/19420889.2015.1119343. eCollection 2016. Commun Integr Biol. 2015. PMID: 27829973 Free PMC article. Review.
-
The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18.J Biol Chem. 2004 Sep 3;279(36):38047-54. doi: 10.1074/jbc.M404319200. Epub 2004 Jun 24. J Biol Chem. 2004. PMID: 15218029
-
The Pam18/Tim14-Pam16/Tim16 complex of the mitochondrial translocation motor: the formation of a stable complex from marginally stable proteins.Protein Sci. 2007 Feb;16(2):316-22. doi: 10.1110/ps.062459607. Protein Sci. 2007. PMID: 17242434 Free PMC article.
-
Reevaluation of the role of the Pam18:Pam16 interaction in translocation of proteins by the mitochondrial Hsp70-based import motor.Mol Biol Cell. 2011 Dec;22(24):4740-9. doi: 10.1091/mbc.E11-08-0715. Epub 2011 Oct 26. Mol Biol Cell. 2011. PMID: 22031295 Free PMC article.
-
The mitochondrial carrier family of transport proteins: structural, functional, and evolutionary relationships.Crit Rev Biochem Mol Biol. 1993;28(3):209-33. doi: 10.3109/10409239309086795. Crit Rev Biochem Mol Biol. 1993. PMID: 8325039 Review.
Cited by
-
J-like protein family of Arabidopsis thaliana: the enigmatic cousins of J-domain proteins.Plant Cell Rep. 2022 Jun;41(6):1343-1355. doi: 10.1007/s00299-022-02857-y. Epub 2022 Mar 15. Plant Cell Rep. 2022. PMID: 35290497 Review.
-
Expansion of the evolutionarily conserved network of J-domain proteins in the Arabidopsis mitochondrial import complex.Plant Mol Biol. 2021 Mar;105(4-5):385-403. doi: 10.1007/s11103-020-01095-8. Epub 2020 Nov 18. Plant Mol Biol. 2021. PMID: 33206359
-
Presequence translocase-associated motor subunits of the mitochondrial protein import apparatus are dual-targeted to mitochondria and plastids.Front Plant Sci. 2022 Nov 11;13:981552. doi: 10.3389/fpls.2022.981552. eCollection 2022. Front Plant Sci. 2022. PMID: 36438081 Free PMC article.
-
Identification of Genomic Structural Variations in Xinjiang Brown Cattle by Deep Sequencing and Their Association with Body Conformation Traits.Int J Mol Sci. 2025 May 29;26(11):5234. doi: 10.3390/ijms26115234. Int J Mol Sci. 2025. PMID: 40508044 Free PMC article.
-
Common functions of the chloroplast and mitochondrial co-chaperones cpDnaJL (CDF1) and mtDnaJ (PAM16) in protein import and ROS scavenging in Arabidopsis thaliana.Commun Integr Biol. 2015 Dec 9;9(5):e1119343. doi: 10.1080/19420889.2015.1119343. eCollection 2016. Commun Integr Biol. 2015. PMID: 27829973 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

