Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States
- PMID: 24197882
- PMCID: PMC3911415
- DOI: 10.1128/JCM.02820-13
Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States
Abstract
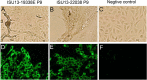
Porcine epidemic diarrhea virus (PEDV) was detected in May 2013 for the first time in U.S. swine and has since caused significant economic loss. Obtaining a U.S. PEDV isolate that can grow efficiently in cell culture is critical for investigating pathogenesis and developing diagnostic assays and for vaccine development. An additional objective was to determine which gene(s) of PEDV is most suitable for studying the genetic relatedness of the virus. Here we describe two PEDV isolates (ISU13-19338E and ISU13-22038) successfully obtained from the small intestines of piglets from sow farms in Indiana and Iowa, respectively. The two isolates have been serially propagated in cell culture for over 30 passages and were characterized for the first 10 passages. Virus production in cell culture was confirmed by PEDV-specific real-time reverse-transcription PCR (RT-PCR), immunofluorescence assays, and electron microscopy. The infectious titers of the viruses during the first 10 passages ranged from 6 × 10(2) to 2 × 10(5) 50% tissue culture infective doses (TCID50)/ml. In addition, the full-length genome sequences of six viruses (ISU13-19338E homogenate, P3, and P9; ISU13-22038 homogenate, P3, and P9) were determined. Genetically, the two PEDV isolates were relatively stable during the first 10 passages in cell culture. Sequences were also compared to those of 4 additional U.S. PEDV strains and 23 non-U.S. strains. All U.S. PEDV strains were genetically closely related to each other (≥99.7% nucleotide identity) and were most genetically similar to Chinese strains reported in 2011 to 2012. Phylogenetic analyses using different genes of PEDV suggested that the full-length spike gene or the S1 portion is appropriate for sequencing to study the genetic relatedness of these viruses.
Figures




Comment in
-
Classification of emergent U.S. strains of porcine epidemic diarrhea virus by phylogenetic analysis of nucleocapsid and ORF3 genes.J Clin Microbiol. 2014 Sep;52(9):3509-10. doi: 10.1128/JCM.01708-14. J Clin Microbiol. 2014. PMID: 25143422 Free PMC article. No abstract available.
-
Reply to "classification of emergent U.S. strains of porcine epidemic diarrhea virus by phylogenetic analysis of nucleocapsid and ORF3 genes".J Clin Microbiol. 2014 Sep;52(9):3511-4. doi: 10.1128/JCM.01747-14. J Clin Microbiol. 2014. PMID: 25143423 Free PMC article. No abstract available.
References
-
- Oldham J. 1972. Letter to the editor. Pig Farming 1972(October suppl):72–73
-
- International Committee on Taxonomy of Viruses 2012. Virus taxonomy: 2012 release. http://ictvonline.org/virusTaxonomy.asp?version=2012
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

