Identification and functional analysis of novel phosphorylation sites in the RNA surveillance protein Upf1
- PMID: 24198248
- PMCID: PMC3919615
- DOI: 10.1093/nar/gkt1049
Identification and functional analysis of novel phosphorylation sites in the RNA surveillance protein Upf1
Abstract
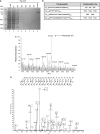
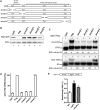
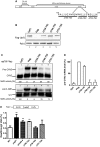
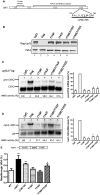
One third of inherited genetic diseases are caused by mRNAs harboring premature termination codons as a result of nonsense mutations. These aberrant mRNAs are degraded by the Nonsense-Mediated mRNA Decay (NMD) pathway. A central component of the NMD pathway is Upf1, an RNA-dependent ATPase and helicase. Upf1 is a known phosphorylated protein, but only portions of this large protein have been examined for phosphorylation sites and the functional relevance of its phosphorylation has not been elucidated in Saccharomyces cerevisiae. Using tandem mass spectrometry analyses, we report the identification of 11 putative phosphorylated sites in S. cerevisiae Upf1. Five of these phosphorylated residues are located within the ATPase and helicase domains and are conserved in higher eukaryotes, suggesting a biological significance for their phosphorylation. Indeed, functional analysis demonstrated that a small carboxy-terminal motif harboring at least three phosphorylated amino acids is important for three Upf1 functions: ATPase activity, NMD activity and the ability to promote translation termination efficiency. We provide evidence that two tyrosines within this phospho-motif (Y-738 and Y-742) act redundantly to promote ATP hydrolysis, NMD efficiency and translation termination fidelity.
Figures


Similar articles
-
Intra- and intermolecular regulatory interactions in Upf1, the RNA helicase central to nonsense-mediated mRNA decay in yeast.Mol Cell Biol. 2013 Dec;33(23):4672-84. doi: 10.1128/MCB.01136-13. Epub 2013 Oct 7. Mol Cell Biol. 2013. PMID: 24100012 Free PMC article.
-
ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons.Nat Commun. 2016 Dec 23;7:14021. doi: 10.1038/ncomms14021. Nat Commun. 2016. PMID: 28008922 Free PMC article.
-
Phosphorylation of the N- and C-terminal UPF1 domains plays a critical role in plant nonsense-mediated mRNA decay.Plant J. 2013 Dec;76(5):836-48. doi: 10.1111/tpj.12346. Epub 2013 Nov 5. Plant J. 2013. PMID: 24118551
-
Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay.Genes Cells. 2013 Mar;18(3):161-75. doi: 10.1111/gtc.12033. Epub 2013 Jan 28. Genes Cells. 2013. PMID: 23356578 Review.
-
Defining nonsense-mediated mRNA decay intermediates in human cells.Methods. 2019 Feb 15;155:68-76. doi: 10.1016/j.ymeth.2018.12.005. Epub 2018 Dec 19. Methods. 2019. PMID: 30576707 Free PMC article. Review.
Cited by
-
Phosphoproteomics technologies and applications in plant biology research.Front Plant Sci. 2015 Jun 16;6:430. doi: 10.3389/fpls.2015.00430. eCollection 2015. Front Plant Sci. 2015. PMID: 26136758 Free PMC article. Review.
-
Characterization of the mIF4G Domains in the RNA Surveillance Protein Upf2p.Curr Issues Mol Biol. 2023 Dec 29;46(1):244-261. doi: 10.3390/cimb46010017. Curr Issues Mol Biol. 2023. PMID: 38248319 Free PMC article.
-
Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story.Annu Rev Genet. 2015;49:339-66. doi: 10.1146/annurev-genet-112414-054639. Epub 2015 Oct 2. Annu Rev Genet. 2015. PMID: 26436458 Free PMC article. Review.
-
Nonsense-mediated mRNA decay involves two distinct Upf1-bound complexes.EMBO J. 2018 Nov 2;37(21):e99278. doi: 10.15252/embj.201899278. Epub 2018 Oct 1. EMBO J. 2018. PMID: 30275269 Free PMC article.
-
Mechanism, factors, and physiological role of nonsense-mediated mRNA decay.Cell Mol Life Sci. 2015 Dec;72(23):4523-44. doi: 10.1007/s00018-015-2017-9. Epub 2015 Aug 18. Cell Mol Life Sci. 2015. PMID: 26283621 Free PMC article. Review.
References
-
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. - PubMed
-
- Amrani N, Dong S, He F, Ganesan R, Ghosh S, Kervestin S, Li C, Mangus DA, Spatrick P, Jacobson A. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 2006;34:39–42. - PubMed
-
- Baker KE, Parker R. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr. Opin. Cell Biol. 2004;16:293–299. - PubMed
-
- Gonzalez CI, Wang W, Peltz SW. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae: a quality control mechanism that degrades transcripts harboring premature termination codons. Cold Spring Harb. Symp. Quant. Biol. 2001;66:321–328. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

