Metabolic reprogramming by viruses in the sunlit and dark ocean
- PMID: 24200126
- PMCID: PMC4053976
- DOI: 10.1186/gb-2013-14-11-r123
Metabolic reprogramming by viruses in the sunlit and dark ocean
Abstract
Background: Marine ecosystem function is largely determined by matter and energy transformations mediated by microbial community interaction networks. Viral infection modulates network properties through mortality, gene transfer and metabolic reprogramming.
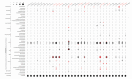
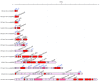
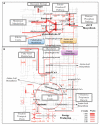
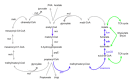
Results: Here we explore the nature and extent of viral metabolic reprogramming throughout the Pacific Ocean depth continuum. We describe 35 marine viral gene families with potential to reprogram metabolic flux through central metabolic pathways recovered from Pacific Ocean waters. Four of these families have been previously reported but 31 are novel. These known and new carbon pathway auxiliary metabolic genes were recovered from a total of 22 viral metagenomes in which viral auxiliary metabolic genes were differentiated from low-level cellular DNA inputs based on small subunit ribosomal RNA gene content, taxonomy, fragment recruitment and genomic context information. Auxiliary metabolic gene distribution patterns reveal that marine viruses target overlapping, but relatively distinct pathways in sunlit and dark ocean waters to redirect host carbon flux towards energy production and viral genome replication under low nutrient, niche-differentiated conditions throughout the depth continuum.
Conclusions: Given half of ocean microbes are infected by viruses at any given time, these findings of broad viral metabolic reprogramming suggest the need for renewed consideration of viruses in global ocean carbon models.
Figures

References
-
- Falkowski PG, Barber RT, Smetacek V. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;14:200–207. - PubMed
-
- Kasting JF, Siefert JL. Life and the evolution of Earth’s atmosphere. Science. 2002;14:1066–1068. - PubMed
-
- Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;14:538–550. - PubMed
-
- Wright JJ, Konwar KM, Hallam SJ. Microbial ecology of expanding oxygen minimum zones. Nat Rev Microbiol. 2012;14:381–394. - PubMed
-
- Breitbart M, Thompson LR, Suttle CA, Sullivan MB. Exploring the vast diversity of marine viruses. Oceanography. 2007;14:135–139.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

