AHT-ChIP-seq: a completely automated robotic protocol for high-throughput chromatin immunoprecipitation
- PMID: 24200198
- PMCID: PMC4053851
- DOI: 10.1186/gb-2013-14-11-r124
AHT-ChIP-seq: a completely automated robotic protocol for high-throughput chromatin immunoprecipitation
Abstract
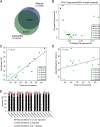
ChIP-seq is an established manually-performed method for identifying DNA-protein interactions genome-wide. Here, we describe a protocol for automated high-throughput (AHT) ChIP-seq. To demonstrate the quality of data obtained using AHT-ChIP-seq, we applied it to five proteins in mouse livers using a single 96-well plate, demonstrating an extremely high degree of qualitative and quantitative reproducibility among biological and technical replicates. We estimated the optimum and minimum recommended cell numbers required to perform AHT-ChIP-seq by running an additional plate using HepG2 and MCF7 cells. With this protocol, commercially available robotics can perform four hundred experiments in five days.
Figures




References
-
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, Bennett DA, Houmard JA, Muoio DM, Onder TT, Camahort R, Cowan CA, Meissner A, Epstein CB, Shoresh N, Bernstein BE. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;14:642–654. doi: 10.1016/j.cell.2012.12.033. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

