RAM function is dependent on Kapβ2-mediated nuclear entry
- PMID: 24200467
- PMCID: PMC3898117
- DOI: 10.1042/BJ20131359
RAM function is dependent on Kapβ2-mediated nuclear entry
Abstract
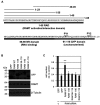
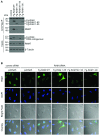
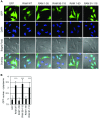
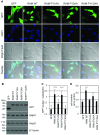
Eukaryotic gene expression is dependent on the modification of the first transcribed nucleotide of pre-mRNA by the addition of the 7-methylguanosine cap. The cap protects transcripts from exonucleases and recruits complexes which mediate transcription elongation, processing and translation initiation. The cap is synthesized by a series of reactions which link 7-methylguanosine to the first transcribed nucleotide via a 5' to 5' triphosphate bridge. In mammals, cap synthesis is catalysed by the sequential action of RNGTT (RNA guanylyltransferase and 5'-phosphatase) and RNMT (RNA guanine-7 methyltransferase), enzymes recruited to RNA pol II (polymerase II) during the early stages of transcription. We recently discovered that the mammalian cap methyltransferase is a heterodimer consisting of RNMT and the RNMT-activating subunit RAM (RNMT-activating mini-protein). RAM activates and stabilizes RNMT and thus is critical for cellular cap methylation and cell viability. In the present study we report that RNMT interacts with the N-terminal 45 amino acids of RAM, a domain necessary and sufficient for maximal RNMT activation. In contrast, smaller components of this RAM domain are sufficient to stabilize RNMT. RAM functions in the nucleus and we report that nuclear import of RAM is dependent on PY nuclear localization signals and Kapβ2 (karyopherin β2) nuclear transport protein.
Figures



References
-
- Topisirovic I., Svitkin Y.V., Sonenberg N., Shatkin A.J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA. 2011;2:277–298. - PubMed
-
- Shatkin A.J. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. - PubMed
-
- Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 2002;3:619–625. - PubMed
-
- Li Y., Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip. Rev. RNA. 2010;1:253–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

