Valvular dystrophy associated filamin A mutations reveal a new role of its first repeats in small-GTPase regulation
- PMID: 24200678
- PMCID: PMC3928473
- DOI: 10.1016/j.bbamcr.2013.10.022
Valvular dystrophy associated filamin A mutations reveal a new role of its first repeats in small-GTPase regulation
Abstract
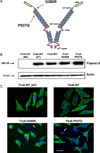
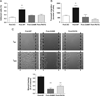
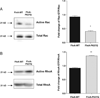
Filamin A (FlnA) is a ubiquitous actin binding protein which anchors various transmembrane proteins to the cell cytoskeleton and provides a scaffold to many cytoplasmic signaling proteins involved in actin cytoskeleton remodeling in response to mechanical stress and cytokines stimulation. Although the vast majority of FlnA binding partners interact with the carboxy-terminal immunoglobulin like (Igl) repeats of FlnA, little is known on the role of the amino-N-terminal repeats. Here, using cardiac mitral valvular dystrophy associated FlnA-G288R and P637Q mutations located in the N-terminal Igl repeat 1 and 4 respectively as a model, we identified a new role of FlnA N-terminal repeats in small Rho-GTPases regulation. Using FlnA-deficient melanoma and HT1080 cell lines as expression systems we showed that FlnA mutations reduce cell spreading and migration capacities. Furthermore, we defined a signaling network in which FlnA mutations alter the balance between RhoA and Rac1 GTPases activities in favor of RhoA and provided evidences for a role of the Rac1 specific GTPase activating protein FilGAP in this process. Together our work ascribed a new role to the N-terminal repeats of FlnA in Small GTPases regulation and supports a conceptual framework for the role of FlnA mutations in cardiac valve diseases centered around signaling molecules regulating cellular actin cytoskeleton in response to mechanical stress.
Keywords: FilGAP; Filamin A; Mitral valve prolapse; Rac1; RhoA.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures




References
-
- Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat. Cell Biol. 2004;6:1034–1038. - PubMed
-
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001;2:138–145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

