Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability
- PMID: 24201445
- PMCID: PMC3912410
- DOI: 10.1101/gr.164806.113
Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability
Abstract
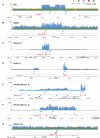
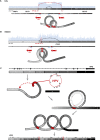
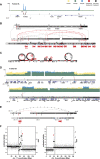
Genomic instability is a hallmark of human cancers, including the 5% caused by human papillomavirus (HPV). Here we report a striking association between HPV integration and adjacent host genomic structural variation in human cancer cell lines and primary tumors. Whole-genome sequencing revealed HPV integrants flanking and bridging extensive host genomic amplifications and rearrangements, including deletions, inversions, and chromosomal translocations. We present a model of "looping" by which HPV integrant-mediated DNA replication and recombination may result in viral-host DNA concatemers, frequently disrupting genes involved in oncogenesis and amplifying HPV oncogenes E6 and E7. Our high-resolution results shed new light on a catastrophic process, distinct from chromothripsis and other mutational processes, by which HPV directly promotes genomic instability.
Figures




References
-
- Ballo H, Koldovsky P, Hoffmann T, Balz V, Hildebrandt B, Gerharz CD, Bier H 1999. Establishment and characterization of four cell lines derived from human head and neck squamous cell carcinomas for an autologous tumor-fibroblast in vitro model. Anticancer Res 19: 3827–3836 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
