RNA editing regulates transposon-mediated heterochromatic gene silencing
- PMID: 24201902
- PMCID: PMC3992701
- DOI: 10.1038/ncomms3745
RNA editing regulates transposon-mediated heterochromatic gene silencing
Abstract
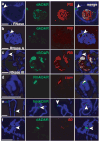
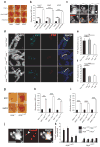
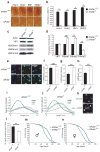
Heterochromatin formation drives epigenetic mechanisms associated with silenced gene expression. Repressive heterochromatin is established through the RNA interference pathway, triggered by double-stranded RNAs (dsRNAs) that can be modified via RNA editing. However, the biological consequences of such modifications remain enigmatic. Here we show that RNA editing regulates heterochromatic gene silencing in Drosophila. We utilize the binding activity of an RNA-editing enzyme to visualize the in vivo production of a long dsRNA trigger mediated by Hoppel transposable elements. Using homologous recombination, we delete this trigger, dramatically altering heterochromatic gene silencing and chromatin architecture. Furthermore, we show that the trigger RNA is edited and that dADAR serves as a key regulator of chromatin state. Additionally, dADAR auto-editing generates a natural suppressor of gene silencing. Lastly, systemic differences in RNA editing activity generates interindividual variation in silencing state within a population. Our data reveal a global role for RNA editing in regulating gene expression.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


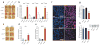
Similar articles
-
dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing.RNA. 2000 Jul;6(7):1004-18. doi: 10.1017/s1355838200000248. RNA. 2000. PMID: 10917596 Free PMC article.
-
Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin.Curr Biol. 2005 Feb 22;15(4):384-91. doi: 10.1016/j.cub.2005.01.046. Curr Biol. 2005. PMID: 15723802
-
Auto-regulatory RNA editing fine-tunes mRNA re-coding and complex behaviour in Drosophila.Nat Commun. 2012 Apr 24;3:790. doi: 10.1038/ncomms1789. Nat Commun. 2012. PMID: 22531175 Free PMC article.
-
How does RNA editing affect dsRNA-mediated gene silencing?Cold Spring Harb Symp Quant Biol. 2006;71:285-92. doi: 10.1101/sqb.2006.71.037. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381308 Free PMC article. Review.
-
RNA silencing in Drosophila.FEBS Lett. 2005 Oct 31;579(26):5940-9. doi: 10.1016/j.febslet.2005.08.069. Epub 2005 Sep 20. FEBS Lett. 2005. PMID: 16198344 Review.
Cited by
-
RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases.Genes (Basel). 2021 Apr 22;12(5):627. doi: 10.3390/genes12050627. Genes (Basel). 2021. PMID: 33922187 Free PMC article. Review.
-
The Research and Applications of Quantum Dots as Nano-Carriers for Targeted Drug Delivery and Cancer Therapy.Nanoscale Res Lett. 2016 Dec;11(1):207. doi: 10.1186/s11671-016-1394-9. Epub 2016 Apr 18. Nanoscale Res Lett. 2016. PMID: 27090658 Free PMC article. Review.
-
Epitranscriptomic profiling across cell types reveals associations between APOBEC1-mediated RNA editing, gene expression outcomes, and cellular function.Proc Natl Acad Sci U S A. 2017 Dec 12;114(50):13296-13301. doi: 10.1073/pnas.1714227114. Epub 2017 Nov 22. Proc Natl Acad Sci U S A. 2017. PMID: 29167373 Free PMC article.
-
Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila.Proc Natl Acad Sci U S A. 2016 Oct 4;113(40):11277-11282. doi: 10.1073/pnas.1604621113. Epub 2016 Sep 12. Proc Natl Acad Sci U S A. 2016. PMID: 27621458 Free PMC article.
-
Cellular labeling of endogenous retrovirus replication (CLEVR) reveals de novo insertions of the gypsy retrotransposable element in cell culture and in both neurons and glial cells of aging fruit flies.PLoS Biol. 2019 May 16;17(5):e3000278. doi: 10.1371/journal.pbio.3000278. eCollection 2019 May. PLoS Biol. 2019. PMID: 31095565 Free PMC article.
References
-
- Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43. - PubMed
-
- Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

