RNA editing regulates transposon-mediated heterochromatic gene silencing
- PMID: 24201902
- PMCID: PMC3992701
- DOI: 10.1038/ncomms3745
RNA editing regulates transposon-mediated heterochromatic gene silencing
Abstract
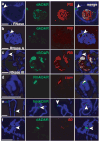
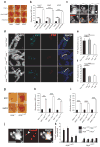
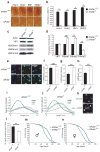
Heterochromatin formation drives epigenetic mechanisms associated with silenced gene expression. Repressive heterochromatin is established through the RNA interference pathway, triggered by double-stranded RNAs (dsRNAs) that can be modified via RNA editing. However, the biological consequences of such modifications remain enigmatic. Here we show that RNA editing regulates heterochromatic gene silencing in Drosophila. We utilize the binding activity of an RNA-editing enzyme to visualize the in vivo production of a long dsRNA trigger mediated by Hoppel transposable elements. Using homologous recombination, we delete this trigger, dramatically altering heterochromatic gene silencing and chromatin architecture. Furthermore, we show that the trigger RNA is edited and that dADAR serves as a key regulator of chromatin state. Additionally, dADAR auto-editing generates a natural suppressor of gene silencing. Lastly, systemic differences in RNA editing activity generates interindividual variation in silencing state within a population. Our data reveal a global role for RNA editing in regulating gene expression.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


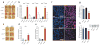
References
-
- Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43. - PubMed
-
- Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

