Synthetic genetic targeting of genome instability in cancer
- PMID: 24202319
- PMCID: PMC3795363
- DOI: 10.3390/cancers5030739
Synthetic genetic targeting of genome instability in cancer
Abstract
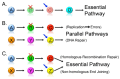
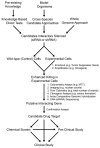
Cancer is a leading cause of death throughout the World. A limitation of many current chemotherapeutic approaches is that their cytotoxic effects are not restricted to cancer cells, and adverse side effects can occur within normal tissues. Consequently, novel strategies are urgently needed to better target cancer cells. As we approach the era of personalized medicine, targeting the specific molecular defect(s) within a given patient's tumor will become a more effective treatment strategy than traditional approaches that often target a given cancer type or sub-type. Synthetic genetic interactions are now being examined for their therapeutic potential and are designed to target the specific genetic and epigenetic phenomena associated with tumor formation, and thus are predicted to be highly selective. In general, two complementary approaches have been employed, including synthetic lethality and synthetic dosage lethality, to target aberrant expression and/or function associated with tumor suppressor genes and oncogenes, respectively. Here we discuss the concepts of synthetic lethality and synthetic dosage lethality, and explain three general experimental approaches designed to identify novel genetic interactors. We present examples and discuss the merits and caveats of each approach. Finally, we provide insight into the subsequent pre-clinical work required to validate novel candidate drug targets.
Figures

Similar articles
-
The enigmatic oncogene and tumor suppressor-like properties of RAD54B: Insights into genome instability and cancer.Genes Chromosomes Cancer. 2017 Jul;56(7):513-523. doi: 10.1002/gcc.22458. Epub 2017 Apr 4. Genes Chromosomes Cancer. 2017. PMID: 28295846 Review.
-
Targeting the CINful genome: Strategies to overcome tumor heterogeneity.Prog Biophys Mol Biol. 2019 Oct;147:77-91. doi: 10.1016/j.pbiomolbio.2019.02.006. Epub 2019 Feb 25. Prog Biophys Mol Biol. 2019. PMID: 30817936 Review.
-
Targeting SOD1 induces synthetic lethal killing in BLM- and CHEK2-deficient colorectal cancer cells.Oncotarget. 2015 Sep 29;6(29):27907-22. doi: 10.18632/oncotarget.4875. Oncotarget. 2015. PMID: 26318585 Free PMC article.
-
Unveiling the vulnerabilities of synthetic lethality in triple-negative breast cancer.Clin Transl Oncol. 2023 Nov;25(11):3057-3072. doi: 10.1007/s12094-023-03191-9. Epub 2023 Apr 20. Clin Transl Oncol. 2023. PMID: 37079210 Review.
-
Advances in Synthetic Lethality in Potential Oncology Therapeutic Approaches.Curr Top Med Chem. 2025 Jan 30. doi: 10.2174/0115680266349547241231051447. Online ahead of print. Curr Top Med Chem. 2025. PMID: 39901548
Cited by
-
Role of RNF20 in cancer development and progression - a comprehensive review.Biosci Rep. 2018 Jul 12;38(4):BSR20171287. doi: 10.1042/BSR20171287. Print 2018 Aug 31. Biosci Rep. 2018. PMID: 29934362 Free PMC article. Review.
-
Aberrant SKP1 Expression: Diverse Mechanisms Impacting Genome and Chromosome Stability.Front Cell Dev Biol. 2022 Mar 8;10:859582. doi: 10.3389/fcell.2022.859582. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35345853 Free PMC article. Review.
-
Development of Metabolic Synthetic Lethality and Its Implications for Thyroid Cancer.Endocrinol Metab (Seoul). 2022 Feb;37(1):53-61. doi: 10.3803/EnM.2022.1402. Epub 2022 Feb 28. Endocrinol Metab (Seoul). 2022. PMID: 35255601 Free PMC article.
-
Predicting chemotherapeutic drug combinations through gene network profiling.Sci Rep. 2016 Jan 21;6:18658. doi: 10.1038/srep18658. Sci Rep. 2016. PMID: 26791325 Free PMC article.
-
Genomic instability in human cancer: Molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition.Semin Cancer Biol. 2015 Dec;35 Suppl(Suppl):S5-S24. doi: 10.1016/j.semcancer.2015.03.005. Epub 2015 Apr 11. Semin Cancer Biol. 2015. PMID: 25869442 Free PMC article. Review.
References
-
- Ferlay J.S.H., Bray F., Forman D., Mathers C., Parkin D.M. International Agency for Research on Cancer; Lyon, France: 2010. Globocan 2008 v2.0, Cancer Incidence And Mortality Worldwide: Iarc Cancerbase No. 10.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

