Src is activated by the nuclear receptor peroxisome proliferator-activated receptor β/δ in ultraviolet radiation-induced skin cancer
- PMID: 24203162
- PMCID: PMC3936491
- DOI: 10.1002/emmm.201302666
Src is activated by the nuclear receptor peroxisome proliferator-activated receptor β/δ in ultraviolet radiation-induced skin cancer
Abstract
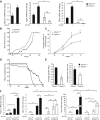
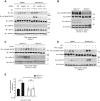
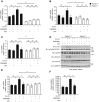
Although non-melanoma skin cancer (NMSC) is the most common human cancer and its incidence continues to rise worldwide, the mechanisms underlying its development remain incompletely understood. Here, we unveil a cascade of events involving peroxisome proliferator-activated receptor (PPAR) β/δ and the oncogene Src, which promotes the development of ultraviolet (UV)-induced skin cancer in mice. UV-induced PPARβ/δ activity, which directly stimulated Src expression, increased Src kinase activity and enhanced the EGFR/Erk1/2 signalling pathway, resulting in increased epithelial-to-mesenchymal transition (EMT) marker expression. Consistent with these observations, PPARβ/δ-null mice developed fewer and smaller skin tumours, and a PPARβ/δ antagonist prevented UV-dependent Src stimulation. Furthermore, the expression of PPARβ/δ positively correlated with the expression of SRC and EMT markers in human skin squamous cell carcinoma (SCC), and critically, linear models applied to several human epithelial cancers revealed an interaction between PPARβ/δ and SRC and TGFβ1 transcriptional levels. Taken together, these observations motivate the future evaluation of PPARβ/δ modulators to attenuate the development of several epithelial cancers.
Figures




References
-
- Adhikary T, Brandt DT, Kaddatz K, Stockert J, Naruhn S, Meissner W, Finkernagel F, Obert J, Lieber S, Scharfe M, et al. Inverse PPARbeta/delta agonists suppress oncogenic signaling to the ANGPTL4 gene and inhibit cancer cell invasion. Oncogene. 2012 DOI: 10.1038/onc.2012.549. - DOI - PMC - PubMed
-
- Allalou A, Wahlby C. BlobFinder, a tool for fluorescence microscopy image cytometry. Comput Methods Programs Biomed. 2009;94:58–65. - PubMed
-
- Bode AM, Dong Z. Mitogen-activated protein kinase activation in UV-induced signal transduction. Sci STKE. 2003;2003:RE2. - PubMed
-
- Boukamp P. Non-melanoma skin cancer: What drives tumor development and progression? Carcinogenesis. 2005;26:1657–1667. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

