Properties of MHC class I presented peptides that enhance immunogenicity
- PMID: 24204222
- PMCID: PMC3808449
- DOI: 10.1371/journal.pcbi.1003266
Properties of MHC class I presented peptides that enhance immunogenicity
Abstract
T-cells have to recognize peptides presented on MHC molecules to be activated and elicit their effector functions. Several studies demonstrate that some peptides are more immunogenic than others and therefore more likely to be T-cell epitopes. We set out to determine which properties cause such differences in immunogenicity. To this end, we collected and analyzed a large set of data describing the immunogenicity of peptides presented on various MHC-I molecules. Two main conclusions could be drawn from this analysis: First, in line with previous observations, we showed that positions P4-6 of a presented peptide are more important for immunogenicity. Second, some amino acids, especially those with large and aromatic side chains, are associated with immunogenicity. This information was combined into a simple model that was used to demonstrate that immunogenicity is, to a certain extent, predictable. This model (made available at http://tools.iedb.org/immunogenicity/) was validated with data from two independent epitope discovery studies. Interestingly, with this model we could show that T-cells are equipped to better recognize viral than human (self) peptides. After the past successful elucidation of different steps in the MHC-I presentation pathway, the identification of variables that influence immunogenicity will be an important next step in the investigation of T-cell epitopes and our understanding of cellular immune responses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

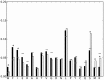
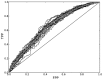
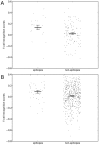

References
-
- Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK (2006) Naive and memory CD4+ T cell survival controlled by clonal abundance. Science 312: 114–116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

