A comprehensive survey of small-molecule binding pockets in proteins
- PMID: 24204237
- PMCID: PMC3812058
- DOI: 10.1371/journal.pcbi.1003302
A comprehensive survey of small-molecule binding pockets in proteins
Abstract
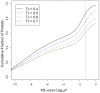
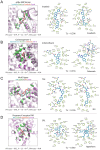
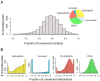
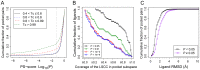
Many biological activities originate from interactions between small-molecule ligands and their protein targets. A detailed structural and physico-chemical characterization of these interactions could significantly deepen our understanding of protein function and facilitate drug design. Here, we present a large-scale study on a non-redundant set of about 20,000 known ligand-binding sites, or pockets, of proteins. We find that the structural space of protein pockets is crowded, likely complete, and may be represented by about 1,000 pocket shapes. Correspondingly, the growth rate of novel pockets deposited in the Protein Data Bank has been decreasing steadily over the recent years. Moreover, many protein pockets are promiscuous and interact with ligands of diverse scaffolds. Conversely, many ligands are promiscuous and interact with structurally different pockets. Through a physico-chemical and structural analysis, we provide insights into understanding both pocket promiscuity and ligand promiscuity. Finally, we discuss the implications of our study for the prediction of protein-ligand interactions based on pocket comparison.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
A new protein binding pocket similarity measure based on comparison of clouds of atoms in 3D: application to ligand prediction.BMC Bioinformatics. 2010 Feb 22;11:99. doi: 10.1186/1471-2105-11-99. BMC Bioinformatics. 2010. PMID: 20175916 Free PMC article.
-
APoc: large-scale identification of similar protein pockets.Bioinformatics. 2013 Mar 1;29(5):597-604. doi: 10.1093/bioinformatics/btt024. Epub 2013 Jan 17. Bioinformatics. 2013. PMID: 23335017 Free PMC article.
-
An Augmented Pocketome: Detection and Analysis of Small-Molecule Binding Pockets in Proteins of Known 3D Structure.Structure. 2018 Mar 6;26(3):499-512.e2. doi: 10.1016/j.str.2018.02.001. Structure. 2018. PMID: 29514079
-
Implications of the small number of distinct ligand binding pockets in proteins for drug discovery, evolution and biochemical function.Bioorg Med Chem Lett. 2015 Mar 15;25(6):1163-70. doi: 10.1016/j.bmcl.2015.01.059. Epub 2015 Feb 3. Bioorg Med Chem Lett. 2015. PMID: 25690787 Free PMC article. Review.
-
[Development and validation of programs for ligand-binding-pocket search].Yakugaku Zasshi. 2011;131(10):1429-35. doi: 10.1248/yakushi.131.1429. Yakugaku Zasshi. 2011. PMID: 21963969 Review. Japanese.
Cited by
-
Strong Ligand-Protein Interactions Derived from Diffuse Ligand Interactions with Loose Binding Sites.Biomed Res Int. 2015;2015:746980. doi: 10.1155/2015/746980. Epub 2015 May 4. Biomed Res Int. 2015. PMID: 26064949 Free PMC article.
-
Current progress and future perspectives of polypharmacology : From the view of non-small cell lung cancer.Semin Cancer Biol. 2021 Jan;68:84-91. doi: 10.1016/j.semcancer.2019.10.019. Epub 2019 Nov 4. Semin Cancer Biol. 2021. PMID: 31698087 Free PMC article. Review.
-
The Recognition of Unrelated Ligands by Identical Proteins.ACS Chem Biol. 2018 Sep 21;13(9):2522-2533. doi: 10.1021/acschembio.8b00443. Epub 2018 Aug 27. ACS Chem Biol. 2018. PMID: 30095890 Free PMC article.
-
Geometric Detection Algorithms for Cavities on Protein Surfaces in Molecular Graphics: A Survey.Comput Graph Forum. 2017 Dec;36(8):643-683. doi: 10.1111/cgf.13158. Epub 2017 Jun 1. Comput Graph Forum. 2017. PMID: 29520122 Free PMC article.
-
Advances in the Development of Shape Similarity Methods and Their Application in Drug Discovery.Front Chem. 2018 Jul 25;6:315. doi: 10.3389/fchem.2018.00315. eCollection 2018. Front Chem. 2018. PMID: 30090808 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

