The deacetylase Sir2 from the yeast Clavispora lusitaniae lacks the evolutionarily conserved capacity to generate subtelomeric heterochromatin
- PMID: 24204326
- PMCID: PMC3814328
- DOI: 10.1371/journal.pgen.1003935
The deacetylase Sir2 from the yeast Clavispora lusitaniae lacks the evolutionarily conserved capacity to generate subtelomeric heterochromatin
Abstract
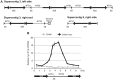
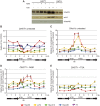
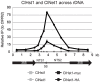
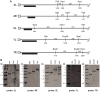
Deacetylases of the Sir2 or sirtuin family are thought to regulate life cycle progression and life span in response to nutrient availability. This family has undergone successive rounds of duplication and diversification, enabling the enzymes to perform a wide variety of biological functions. Two evolutionarily conserved functions of yeast Sir2 proteins are the generation of repressive chromatin in subtelomeric domains and the suppression of unbalanced recombination within the tandem rDNA array. Here, we describe the function of the Sir2 ortholog ClHst1 in the yeast Clavispora lusitaniae, an occasional opportunistic pathogen. ClHst1 was localized to the non-transcribed spacer regions of the rDNA repeats and deacetylated histones at these loci, indicating that, like other Sir2 proteins, ClHst1 modulates chromatin structure at the rDNA repeats. However, we found no evidence that ClHst1 associates with subtelomeric regions or impacts gene expression directly. This surprising observation highlights the plasticity of sirtuin function. Related yeast species, including Candida albicans, possess an additional Sir2 family member. Thus, it is likely that the ancestral Candida SIR2/HST1 gene was duplicated and subfunctionalized, such that HST1 retained the capacity to regulate rDNA whereas SIR2 had other functions, perhaps including the generation of subtelomeric chromatin. After subsequent species diversification, the SIR2 paralog was apparently lost in the C. lusitaniae lineage. Thus, C. lusitaniae presents an opportunity to discover how subtelomeric chromatin can be reconfigured.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Frye RA (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273: 793–798. - PubMed
-
- Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800. - PubMed
-
- Jackson MD, Denu JM (2002) Structural identification of 2′- and 3′-O-acetyl-ADP ribose as novel metabolites derived from the Sir2 family of beta-NAD+-dependant histone/protein deacetylases. J Biol Chem 277: 18535–18544. - PubMed
-
- Guarente L (2000) Sir2 links chromatin silencing, metabolism, and aging. Genes Dev 14: 1021–1026. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

