A method for selectively enriching microbial DNA from contaminating vertebrate host DNA
- PMID: 24204593
- PMCID: PMC3810253
- DOI: 10.1371/journal.pone.0076096
A method for selectively enriching microbial DNA from contaminating vertebrate host DNA
Abstract
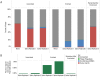
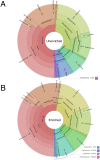
DNA samples derived from vertebrate skin, bodily cavities and body fluids contain both host and microbial DNA; the latter often present as a minor component. Consequently, DNA sequencing of a microbiome sample frequently yields reads originating from the microbe(s) of interest, but with a vast excess of host genome-derived reads. In this study, we used a methyl-CpG binding domain (MBD) to separate methylated host DNA from microbial DNA based on differences in CpG methylation density. MBD fused to the Fc region of a human antibody (MBD-Fc) binds strongly to protein A paramagnetic beads, forming an effective one-step enrichment complex that was used to remove human or fish host DNA from bacterial and protistan DNA for subsequent sequencing and analysis. We report enrichment of DNA samples from human saliva, human blood, a mock malaria-infected blood sample and a black molly fish. When reads were mapped to reference genomes, sequence reads aligning to host genomes decreased 50-fold, while bacterial and Plasmodium DNA sequences reads increased 8-11.5-fold. The Shannon-Wiener diversity index was calculated for 149 bacterial species in saliva before and after enrichment. Unenriched saliva had an index of 4.72, while the enriched sample had an index of 4.80. The similarity of these indices demonstrates that bacterial species diversity and relative phylotype abundance remain conserved in enriched samples. Enrichment using the MBD-Fc method holds promise for targeted microbiome sequence analysis across a broad range of sample types.
Conflict of interest statement
Figures







References
-
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI (2005) Host-bacterial mutualism in the human intestine. Science (New York, NY) 307: 1915–1920. Available: http://www.ncbi.nlm.nih.gov/pubmed/15790844. Accessed 23 May 2013. - PubMed
-
- Fujimura KE, Slusher NA, Cabana MD, Lynch SV (2010) Role of the gut microbiota in defining human health. Expert Review of Anti-infective Therapy 8: 435–454. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2881665&tool=p.... Accessed 20 June 2013. - PMC - PubMed
-
- Koren O, Spor A, Felin J, Fåk F, Stombaugh J, et al.. (2011) Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proceedings of the National Academy of Sciences of the United States of America 108 Suppl: 4592–4598. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3063583&tool=p.... Accessed 26 May 2013. - PMC - PubMed
-
- Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, et al.. (2012) Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PloS One 7: e37919. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3366996&tool=p.... Accessed 27 May 2013. - PMC - PubMed
-
- Kinross JM, Darzi AW, Nicholson JK (2011) Gut microbiome-host interactions in health and disease. Genome Medicine 3: 14. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3092099&tool=p.... Accessed 16 July 2013. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

