Role of microRNA 1207-5P and its host gene, the long non-coding RNA Pvt1, as mediators of extracellular matrix accumulation in the kidney: implications for diabetic nephropathy
- PMID: 24204837
- PMCID: PMC3808414
- DOI: 10.1371/journal.pone.0077468
Role of microRNA 1207-5P and its host gene, the long non-coding RNA Pvt1, as mediators of extracellular matrix accumulation in the kidney: implications for diabetic nephropathy
Erratum in
-
Correction: Role of MicroRNA 1207-5P and Its Host Gene, the Long Non-Coding RNA Pvt1, as Mediators of Extracellular Matrix Accumulation in the Kidney: Implications for Diabetic Nephropathy.PLoS One. 2016 Dec 9;11(12):e0168353. doi: 10.1371/journal.pone.0168353. eCollection 2016. PLoS One. 2016. PMID: 27936176 Free PMC article.
Abstract
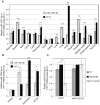
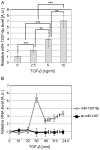
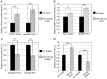
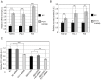
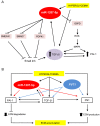
Diabetic nephropathy is the most common cause of chronic kidney failure and end-stage renal disease in the Western World. One of the major characteristics of this disease is the excessive accumulation of extracellular matrix (ECM) in the kidney glomeruli. While both environmental and genetic determinants are recognized for their role in the development of diabetic nephropathy, epigenetic factors, such as DNA methylation, long non-coding RNAs, and microRNAs, have also recently been found to underlie some of the biological mechanisms, including ECM accumulation, leading to the disease. We previously found that a long non-coding RNA, the plasmacytoma variant translocation 1 (PVT1), increases plasminogen activator inhibitor 1 (PAI-1) and transforming growth factor beta 1 (TGF-β1) in mesangial cells, the two main contributors to ECM accumulation in the glomeruli under hyperglycemic conditions, as well as fibronectin 1 (FN1), a major ECM component. Here, we report that miR-1207-5p, a PVT1-derived microRNA, is abundantly expressed in kidney cells, and is upregulated by glucose and TGF-β1. We also found that like PVT1, miR-1207-5p increases expression of TGF-β1, PAI-1, and FN1 but in a manner that is independent of its host gene. In addition, regulation of miR-1207-5p expression by glucose and TGFβ1 is independent of PVT1. These results provide evidence supporting important roles for miR-1207-5p and its host gene in the complex pathogenesis of diabetic nephropathy.
Conflict of interest statement
Figures


References
-
- Kanwar YS, Wada J, Sun L, Xie P, Wallner EI, et al. (2008) Diabetic nephropathy: mechanisms of renal disease progression. Exp Biol Med 233: 4–11. - PubMed
-
- Nelson RG, Knowler WC, Pettitt DJ, Hanson RL, Bennett PH, et al. (1995) Incidence and determinants of elevated urinary albumin excretion in Pima Indians with NIDDM. Diabetes Care 18: 182–187. - PubMed
-
- Ravid M, Brosh D, Ravid-Safran D, Levy Z, Rachmani R (1998) Main risk factors for nephropathy in type 2 diabetes mellitus are plasma cholesterol levels, mean blood pressure, and hyperglycemia. Arch Intern Med 158: 998–1004. - PubMed
-
- Thomas MC, Groop PH, Tryggvason K (2012) Towards understanding the inherited susceptibility for nephropathy in diabetes. Curr Opin Nephrol Hypertens 21: 195–202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

