Characterization of a novel polyomavirus isolated from a fibroma on the trunk of an African elephant (Loxodonta africana)
- PMID: 24205012
- PMCID: PMC3799753
- DOI: 10.1371/journal.pone.0077884
Characterization of a novel polyomavirus isolated from a fibroma on the trunk of an African elephant (Loxodonta africana)
Abstract
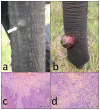
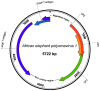
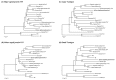
Viruses of the family Polyomaviridae infect a wide variety of avian and mammalian hosts with a broad spectrum of outcomes including asymptomatic infection, acute systemic disease, and tumor induction. In this study a novel polyomavirus, the African elephant polyomavirus 1 (AelPyV-1) found in a protruding hyperplastic fibrous lesion on the trunk of an African elephant (Loxodonta africana) was characterized. The AelPyV-1 genome is 5722 bp in size and is one of the largest polyomaviruses characterized to date. Analysis of the AelPyV-1 genome reveals five putative open-reading frames coding for the classic small and large T antigens in the early region, and the VP1, VP2 and VP3 capsid proteins in the late region. In the area preceding the VP2 start codon three putative open-reading frames, possibly coding for an agnoprotein, could be localized. A regulatory, non-coding region separates the 2 coding regions. Unique for polyomaviruses is the presence of a second 854 bp long non-coding region between the end of the early region and the end of the late region. Based on maximum likelihood phylogenetic analyses of the large T antigen of the AelPyV-1 and 61 other polyomavirus sequences, AelPyV-1 clusters within a heterogeneous group of polyomaviruses that have been isolated from bats, new world primates and rodents.
Conflict of interest statement
Figures

References
-
- Imperiale MJ, Major EO (2007) Polyomaviruses. In: Knipe DM, Howley PM. Fields Virology. Philadelpia. Lippincott Williams and Wilkins; . pp. 2263-2298
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

