Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes
- PMID: 24205083
- PMCID: PMC3804553
- DOI: 10.1371/journal.pone.0078045
Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes
Abstract
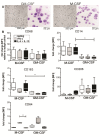
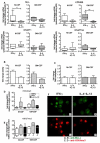
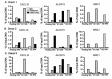
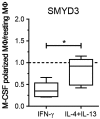
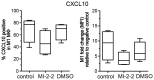
Macrophages (MΦ) play an essential role in innate immune responses and can either display a pro-inflammatory, classically activated phenotype (M1) or undergo an alternative activation program (M2) promoting immune regulation. M-CSF is used to differentiate monocytes into MΦ and IFN-γ or IL-4+IL-13 to further polarize these cells towards M1 or M2, respectively. Recently, differentiation using only GM-CSF or M-CSF has been described to induce a M1- or M2-like phenotype, respectively. In this study, we combined both approaches by differentiating human MΦ in GM-CSF or M-CSF followed by polarization with either IFN-γ or IL-4+IL-13. We describe the phenotypic differences between CD14(hi) CD163(hi) CD206(int) FOLR2-expressing M-CSF MΦ and CD14(lo) CD163(lo) CD206(hi) GM-CSF MΦ but show that both macrophage populations reacted similarly to further polarization with IFN-γ or IL-4+IL-13 with up- and down-regulation of common M1 and M2 marker genes. We also show that high expression of the mannose receptor (CD206), a marker of alternative activation, is a distinct feature of GM-CSF MΦ. Changes of the chromatin structure carried out by chromatin modification enzymes (CME) have been shown to regulate myeloid differentiation. We analyzed the expression patterns of CME during MΦ polarization and show that M1 up-regulate the histone methyltransferase MLL and demethylase KDM6B, while resting and M2 MΦ were characterized by DNA methyltransferases and histone deacetylases. We demonstrate that MLL regulates CXCL10 expression and that this effect could be abrogated using a MLL-Menin inhibitor. Taken together we describe the distinct phenotypic differences of GM-CSF or M-CSF MΦ and demonstrate that MΦ polarization is regulated by specific epigenetic mechanisms. In addition, we describe a novel role for MLL as marker for classical activation. Our findings provide new insights into MΦ polarization that could be helpful to distinguish MΦ activation states.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

