Whole exome sequencing identifies a troponin T mutation hot spot in familial dilated cardiomyopathy
- PMID: 24205113
- PMCID: PMC3812167
- DOI: 10.1371/journal.pone.0078104
Whole exome sequencing identifies a troponin T mutation hot spot in familial dilated cardiomyopathy
Abstract
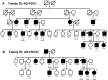
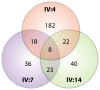
Dilated cardiomyopathy (DCM) commonly causes heart failure and shows extensive genetic heterogeneity that may be amenable to newly developed next-generation DNA sequencing of the exome. In this study we report the successful use of exome sequencing to identify a pathogenic variant in the TNNT2 gene using segregation analysis in a large DCM family. Exome sequencing was performed on three distant relatives from a large family with a clear DCM phenotype. Missense, nonsense, and splice variants were analyzed for segregation among the three affected family members and confirmed in other relatives by direct sequencing. A c.517T C>T, Arg173Trp TNNT2 variant segregated with all affected family members and was also detected in one additional DCM family in our registry. The inclusion of segregation analysis using distant family members markedly improved the bioinformatics filtering process by removing from consideration variants that were not shared by all affected subjects. Haplotype analysis confirmed that the variant found in both DCM families was located on two distinct haplotypes, supporting the notion of independent mutational events in each family. In conclusion, an exome sequencing strategy that includes segregation analysis using distant affected relatives within a family represents a viable diagnostic strategy in a genetically heterogeneous disease like DCM.
Conflict of interest statement
Figures

References
-
- Codd MB, Sugrue DD, Gersh BJ, Melton LJ (1989) Epidemiology of idiopathic dilated and hypertrophic cardiomyopathy. A population-based study in olmsted county, minnesota, 1975–1984. Circulation. 80: 564–572. - PubMed
-
- Grünig E, Tasman JA, Kucherer H, Franz W, Kubler W, et al. (1998) Frequency and phenotypes of familial dilated cardiomyopathy. J Am Coll Cardiol. 31: 186–194. - PubMed
-
- Jefferies JL, Towbin JA (2010) Dilated cardiomyopathy. Lancet. 375: 752–762. - PubMed
-
- Michels VV, Moll PP, Miller FA, Tajik AJ, Chu JS, et al. (1992) The frequency of familial dilated cardiomyopathy in a series of patients with idiopathic dilated cardiomyopathy. N Engl J Med. 326: 77–82. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

