Global Population Structure of the Genes Encoding the Malaria Vaccine Candidate, Plasmodium vivax Apical Membrane Antigen 1 (PvAMA1)
- PMID: 24205419
- PMCID: PMC3814406
- DOI: 10.1371/journal.pntd.0002506
Global Population Structure of the Genes Encoding the Malaria Vaccine Candidate, Plasmodium vivax Apical Membrane Antigen 1 (PvAMA1)
Abstract
Background: The Plasmodium vivax Apical Membrane Antigen 1 (PvAMA1) is a promising malaria vaccine candidate, however it remains unclear which regions are naturally targeted by host immunity and whether its high genetic diversity will preclude coverage by a monovalent vaccine. To assess its feasibility as a vaccine candidate, we investigated the global population structure of PvAMA1.
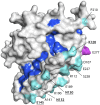
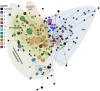
Methodology and principal findings: New sequences from Papua New Guinea (PNG, n = 102) were analysed together with published sequences from Thailand (n = 158), India (n = 8), Sri Lanka (n = 23), Venezuela (n = 74) and a collection of isolates from disparate geographic locations (n = 8). A total of 92 single nucleotide polymorphisms (SNPs) were identified including 22 synonymous SNPs and 70 non-synonymous (NS) SNPs. Polymorphisms and signatures of balancing (positive Tajima's D and low FST values) selection were predominantly clustered in domain I, suggesting it is a dominant target of protective immune responses. To estimate global antigenic diversity, haplotypes comprised of (i) non-singleton (n = 40) and (ii) common (≥10% minor allele frequency, n = 23) polymorphic amino acid sites were then analysed revealing a total of 219 and 210 distinct haplotypes, respectively. Although highly diverse, the 210 haplotypes comprised of only common polymorphisms were grouped into eleven clusters, however substantial geographic differentiation was observed, and this may have implications for the efficacy of PvAMA1 vaccines in different malaria-endemic areas. The PNG haplotypes form a distinct group of clusters not found in any other geographic region. Vaccine haplotypes were rare and geographically restricted, suggesting potentially poor efficacy of candidate PvAMA1 vaccines.
Conclusions: It may be possible to cover the existing global PvAMA1 diversity by selection of diverse alleles based on these analyses however it will be important to first define the relationships between the genetic and antigenic diversity of this molecule.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, et al. (2009) Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis 9: 555–566. - PubMed
-
- Feachem RGA, with A.A Phillips and G.A Targett (eds) (2009) Shrinking the Malaria Map: A Prospectus on Malaria Elimination. San Francisco: The Global Health Group, Global Health Sciences, University of California, San Francisco.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

