TGF-β regulation of gene expression at early and late stages of HPV16-mediated transformation of human keratinocytes
- PMID: 24210100
- PMCID: PMC3895483
- DOI: 10.1016/j.virol.2013.08.034
TGF-β regulation of gene expression at early and late stages of HPV16-mediated transformation of human keratinocytes
Abstract
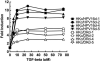
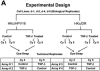
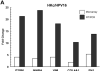
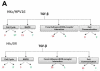
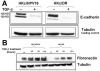
In our in vitro model for HPV16-mediated transformation, HPV16-immortalized human keratinocytes (HKc/HPV16) give rise to differentiation resistant, premalignant cells (HKc/DR). HKc/DR, but not HKc/HPV16, are resistant to growth inhibition by transforming growth factor beta (TGF-β), due to a partial loss of TGF-β receptor type I. We show that TGF-β activates a Smad-responsive reporter construct in HKc/DR to about 50% of the maximum levels of activation observed in HKc/HPV16. To investigate the functional significance of residual TGF-β signaling in HKc/DR, we compared gene expression profiles elicited by TGF-β treatment of HKc/HPV16 and HKc/DR on Agilent 44k human whole genome microarrays. TGF-β altered the expression of cell cycle and MAP kinase pathway genes in HKc/HPV16, but not in HKc/DR. However, epithelial-mesenchymal transition (EMT) responses to TGF-β were comparable in HKc/HPV16 and HKc/DR, indicating that the signaling pathways through which TGF-β elicits growth inhibition diverge from those that induce EMT in HPV16-transformed cells.
Keywords: EMT; HPV; Human keratinocytes; Ski; Smad; TGF-beta.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures








Similar articles
-
Six1 promotes epithelial-mesenchymal transition and malignant conversion in human papillomavirus type 16-immortalized human keratinocytes.Carcinogenesis. 2014 Jun;35(6):1379-88. doi: 10.1093/carcin/bgu050. Epub 2014 Feb 26. Carcinogenesis. 2014. PMID: 24574515 Free PMC article.
-
Partial loss of Smad signaling during in vitro progression of HPV16-immortalized human keratinocytes.BMC Cancer. 2013 Sep 18;13:424. doi: 10.1186/1471-2407-13-424. BMC Cancer. 2013. PMID: 24047375 Free PMC article.
-
Loss of transforming growth factor-beta (TGF-beta) receptor type I mediates TGF-beta resistance in human papillomavirus type 16-transformed human keratinocytes at late stages of in vitro progression.Virology. 2000 May 10;270(2):408-16. doi: 10.1006/viro.2000.0283. Virology. 2000. PMID: 10793000
-
[HPV16 participates in progressive transformation of normal epidermal cells].Gan To Kagaku Ryoho. 1989 Mar;16(3 Pt 2):549-61. Gan To Kagaku Ryoho. 1989. PMID: 2539784 Review. Japanese.
-
Viruses as key modulators of the TGF-β pathway; a double-edged sword involved in cancer.Rev Med Virol. 2018 Mar;28(2):e1967. doi: 10.1002/rmv.1967. Epub 2018 Jan 18. Rev Med Virol. 2018. PMID: 29345394 Free PMC article. Review.
Cited by
-
The translational significance of epithelial-mesenchymal transition in head and neck cancer.Clin Transl Med. 2014 Nov 30;3(1):60. doi: 10.1186/s40169-014-0039-9. eCollection 2014 Dec. Clin Transl Med. 2014. PMID: 25632320 Free PMC article.
-
The Interplay between Antiviral Signalling and Carcinogenesis in Human Papillomavirus Infections.Cancers (Basel). 2020 Mar 10;12(3):646. doi: 10.3390/cancers12030646. Cancers (Basel). 2020. PMID: 32164347 Free PMC article. Review.
-
Induction of Interferon Kappa in Human Papillomavirus 16 Infection by Transforming Growth Factor Beta-Induced Promoter Demethylation.J Virol. 2018 Mar 28;92(8):e01714-17. doi: 10.1128/JVI.01714-17. Print 2018 Apr 15. J Virol. 2018. PMID: 29437968 Free PMC article.
-
Identification of Deregulated Pathways, Key Regulators, and Novel miRNA-mRNA Interactions in HPV-Mediated Transformation.Cancers (Basel). 2020 Mar 16;12(3):700. doi: 10.3390/cancers12030700. Cancers (Basel). 2020. PMID: 32188026 Free PMC article.
-
Stem Cell Properties of Normal Human Keratinocytes Determine Transformation Responses to Human Papillomavirus 16 DNA.J Virol. 2018 May 14;92(11):e00331-18. doi: 10.1128/JVI.00331-18. Print 2018 Jun 1. J Virol. 2018. PMID: 29593030 Free PMC article.
References
-
- Batova A, Danielpour D, Pirisi L, Creek KE. Retinoic acid induces secretion of latent transforming growth factor beta 1 and beta 2 in normal and human papillomavirus type 16-immortalized human keratinocytes. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research. 1992;3:763–772. - PubMed
-
- Bernat A, Avvakumov N, Mymryk JS, Banks L. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene. 2003;22:7871–7881. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

