DUB-resistant ubiquitin to survey ubiquitination switches in mammalian cells
- PMID: 24210823
- PMCID: PMC3889155
- DOI: 10.1016/j.celrep.2013.10.008
DUB-resistant ubiquitin to survey ubiquitination switches in mammalian cells
Abstract
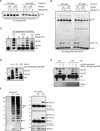
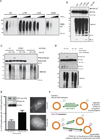
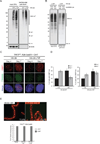
The ubiquitin-modification status of proteins in cells is highly dynamic and maintained by specific ligation machineries (E3 ligases) that tag proteins with ubiquitin or by deubiquitinating enzymes (DUBs) that remove the ubiquitin tag. The development of tools that offset this balance is critical in characterizing signaling pathways that utilize such ubiquitination switches. Herein, we generated a DUB-resistant ubiquitin mutant that is recalcitrant to cleavage by various families of DUBs both in vitro and in mammalian cells. As a proof-of-principle experiment, ectopic expression of the uncleavable ubiquitin stabilized monoubiquitinated PCNA in the absence of DNA damage and also revealed a defect in the clearance of the DNA damage response at unprotected telomeres. Importantly, a proteomic survey using the uncleavable ubiquitin identified ubiquitinated substrates, validating the DUB-resistant ubiquitin expression system as a valuable tool for interrogating cell signaling pathways.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of Interest
B.B. and F.M. are employees of Boston Biochem, Inc. The rest of the authors declare no conflicts of interest.
Figures


References
-
- Bohren KM, Gabbay KH, Owerbach D. Affinity chromatography of native SUMO proteins using His-tagged recombinant UBC9 bound to Co2+-charged talon resin. Protein Expr Purif. 2007;54:289–294. - PubMed
-
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

