Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance
- PMID: 24210919
- PMCID: PMC3974623
- DOI: 10.1016/j.cell.2013.10.027
Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance
Abstract
The regulation of protein-coding and noncoding RNAs is linked to nuclear processes, including chromatin modifications and gene silencing. However, the mechanisms that distinguish RNAs and mediate their functions are poorly understood. We describe a nuclear RNA-processing network in fission yeast with a core module comprising the Mtr4-like protein, Mtl1, and the zinc-finger protein, Red1. The Mtl1-Red1 core promotes degradation of mRNAs and noncoding RNAs and associates with different proteins to assemble heterochromatin via distinct mechanisms. Mtl1 also forms Red1-independent interactions with evolutionarily conserved proteins named Nrl1 and Ctr1, which associate with splicing factors. Whereas Nrl1 targets transcripts with cryptic introns to form heterochromatin at developmental genes and retrotransposons, Ctr1 functions in processing intron-containing telomerase RNA. Together with our discovery of widespread cryptic introns, including in noncoding RNAs, these findings reveal unique cellular strategies for recognizing regulatory RNAs and coordinating their functions in response to developmental and environmental cues.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

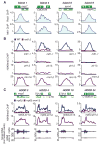
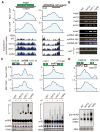


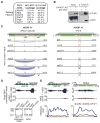

References
-
- Averbeck N, Sunder S, Sample N, Wise JA, Leatherwood J. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol Cell. 2005;18:491–498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

