C(m)CGG methylation-independent parent-of-origin effects on genome-wide transcript levels in isogenic reciprocal F1 triploid plants
- PMID: 24212467
- PMCID: PMC3989486
- DOI: 10.1093/dnares/dst046
C(m)CGG methylation-independent parent-of-origin effects on genome-wide transcript levels in isogenic reciprocal F1 triploid plants
Abstract
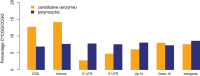
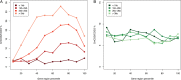
Triploid F1 hybrids generated via reciprocal interploidy crosses between genetically distinct parental plants can display parent-of-origin effects on gene expression or phenotypes. Reciprocal triploid F1 isogenic plants generated from interploidy crosses in the same genetic background allow investigation on parent-of-origin-specific (parental) genome-dosage effects without confounding effects of hybridity involving heterozygous mutations. Whole-genome transcriptome profiling was conducted on reciprocal F1 isogenic triploid (3x) seedlings of A. thaliana. The genetically identical reciprocal 3x genotypes had either an excess of maternally inherited 3x(m) or paternally inherited 3x(p) genomes. We identify a major parent-of-origin-dependent genome-dosage effect on transcript levels, whereby 602 genes exhibit differential expression between the reciprocal F1 triploids. In addition, using methylation-sensitive DNA tiling arrays, constitutive and polymorphic CG DNA methylation patterns at CCGG sites were analysed, which revealed that paternal-excess F1 triploid seedling C(m)CGG sites are overall hypermethylated. However, no correlation exists between C(m)CGG methylation polymorphisms and transcriptome dysregulation between the isogenic reciprocal F1 triploids. Overall, our study indicates that parental genome-dosage effects on the transcriptome levels occur in paternal-excess triploids, which are independent of C(m)CGG methylation polymorphisms. Such findings have implications for understanding parental effects and genome-dosage effects on gene expression and phenotypes in polyploid plants.
Keywords: DNA methylation; epigenetic; parent-of-origin effect; polyploidy; triploid.
Figures


References
-
- Jiao Y., Wickett N.J., Ayyampalayam S., et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. - PubMed
-
- Madlung A., Tyagi A.P., Watson B., et al. Genomic changes in synthetic A. thaliana polyploids. Plant J. 2005;41:221–30. - PubMed
-
- Verhoeven K.J., Van Dijk P.J., Biere A. Changes in genomic methylation patterns during the formation of triploid asexual dandelion lineages. Mol. Ecol. 2010;19:315–24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

