Genome-guided discovery of diverse natural products from Burkholderia sp
- PMID: 24212473
- PMCID: PMC3946939
- DOI: 10.1007/s10295-013-1376-1
Genome-guided discovery of diverse natural products from Burkholderia sp
Abstract
Burkholderia species have emerged as a new source of diverse natural products. This mini-review covers all of the natural products discovered in recent years from Burkholderia sp. by genome-guided approaches--these refer to the use of bacterial genome sequence as an entry point for in silico structural prediction, wet lab experimental design, and execution. While reliable structural prediction based on cryptic biosynthetic gene cluster sequence was not always possible due to noncanonical domains and/or module organization of a deduced biosynthetic pathway, a molecular genetic method was often employed to detect or alter the expression level of the gene cluster to achieve an observable phenotype, which facilitated downstream natural product purification and identification. Those examples of natural product discovery from Burkholderia sp. provide practical guidance for future exploration of Gram-negative bacteria as a new source of natural products.
Figures





References
-
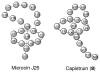
- Bayro MJ, Mukhopadhyay J, Swapna GV, Huang JY, Ma LC, Sineva E, Dawson PE, Montelione GT, Ebright RH. Structure of antibacterial peptide microcin J25: a 21-residue lariat protoknot. J Am Chem Soc. 2003;125:12382–12383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

