Aberrant signaling pathways in glioma
- PMID: 24212955
- PMCID: PMC3759196
- DOI: 10.3390/cancers3033242
Aberrant signaling pathways in glioma
Abstract
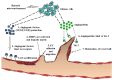
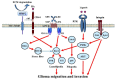
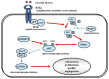
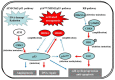
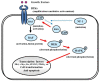
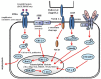
Glioblastoma multiforme (GBM), a WHO grade IV malignant glioma, is the most common and lethal primary brain tumor in adults; few treatments are available. Median survival rates range from 12-15 months. The biological characteristics of this tumor are exemplified by prominent proliferation, active invasiveness, and rich angiogenesis. This is mainly due to highly deregulated signaling pathways in the tumor. Studies of these signaling pathways have greatly increased our understanding of the biology and clinical behavior of GBM. An integrated view of signal transduction will provide a more useful approach in designing novel therapies for this devastating disease. In this review, we summarize the current understanding of GBM signaling pathways with a focus on potential molecular targets for anti-signaling molecular therapies.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

