Nuclear lamins: making contacts with promoters
- PMID: 24213377
- PMCID: PMC3925686
- DOI: 10.4161/nucl.26865
Nuclear lamins: making contacts with promoters
Abstract
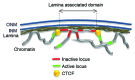
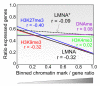
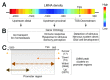
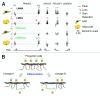
The nuclear lamina guards the genome and in many ways contributes to regulating nuclear function. Increasing evidence indicates that the lamina dynamically interacts with chromatin mainly through large repressive domains, and recent data suggest that at least some of the lamin-genome contacts may be developmentally significant. In an attempt to provide an additional meaning to lamin-genome contacts, a recent study characterized the association of gene promoters with A-type lamins in progenitor and differentiated cells. Here, we discuss how A-type lamins interact with spatially defined promoter regions, and the relationship between these interactions, associated chromatin marks and gene expression outputs. We discuss the impact of A-type lamins on nucleus-wide and local chromatin organization. We also address how lamin-promoter interactions are redistributed during differentiation of adipocyte progenitors into adipocytes. Finally, we propose a model of lineage-specific "unlocking" of developmentally regulated loci and its significance in cellular differentiation.
Keywords: chromatin; differentiation; lamin A/C; lamina-associated domain; nuclear lamina; promoter.
Figures


Comment on
-
Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes.Genome Res. 2013 Oct;23(10):1580-9. doi: 10.1101/gr.159400.113. Epub 2013 Jul 16. Genome Res. 2013. PMID: 23861385 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
