Discovery of recurrent structural variants in nasopharyngeal carcinoma
- PMID: 24214394
- PMCID: PMC3912420
- DOI: 10.1101/gr.156224.113
Discovery of recurrent structural variants in nasopharyngeal carcinoma
Abstract
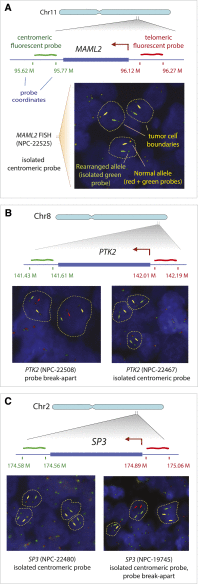
We present the discovery of genes recurrently involved in structural variation in nasopharyngeal carcinoma (NPC) and the identification of a novel type of somatic structural variant. We identified the variants with high complexity mate-pair libraries and a novel computational algorithm specifically designed for tumor-normal comparisons, SMASH. SMASH combines signals from split reads and mate-pair discordance to detect somatic structural variants. We demonstrate a >90% validation rate and a breakpoint reconstruction accuracy of 3 bp by Sanger sequencing. Our approach identified three in-frame gene fusions (YAP1-MAML2, PTPLB-RSRC1, and SP3-PTK2) that had strong levels of expression in corresponding NPC tissues. We found two cases of a novel type of structural variant, which we call "coupled inversion," one of which produced the YAP1-MAML2 fusion. To investigate whether the identified fusion genes are recurrent, we performed fluorescent in situ hybridization (FISH) to screen 196 independent NPC cases. We observed recurrent rearrangements of MAML2 (three cases), PTK2 (six cases), and SP3 (two cases), corresponding to a combined rate of structural variation recurrence of 6% among tested NPC tissues.
Figures




References
-
- Battifora H 1986. The multitumor (sausage) tissue block: Novel method for immunohistochemical antibody testing. Lab Invest 55: 244–249 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
