The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs
- PMID: 24214967
- PMCID: PMC3919597
- DOI: 10.1093/nar/gkt1050
The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs
Abstract
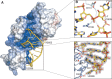
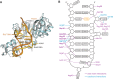
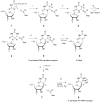
RluB catalyses the modification of U2605 to pseudouridine (Ψ) in a stem-loop at the peptidyl transferase center of Escherichia coli 23S rRNA. The homolog RluF is specific to the adjacent nucleotide in the stem, U2604. The 1.3 Å resolution crystal structure of the complex between the catalytic domain of RluB and the isolated substrate stem-loop, in which the target uridine is substituted by 5-fluorouridine (5-FU), reveals a covalent bond between the isomerized target base and tyrosine 140. The structure is compared with the catalytic domain alone determined at 2.5 Å resolution. The RluB-bound stem-loop has essentially the same secondary structure as in the ribosome, with a bulge at A2602, but with 5-FU2605 flipped into the active site. We showed earlier that RluF induced a frame-shift of the RNA, moving A2602 into the stem and translating its target, U2604, into the active site. A hydrogen-bonding network stabilizes the bulge in the RluB-RNA but is not conserved in RluF and so RluF cannot stabilize the bulge. On the basis of the covalent bond between enzyme and isomerized 5-FU we propose a Michael addition mechanism for pseudouridine formation that is consistent with all experimental data.
Figures




Similar articles
-
Crystal structure of an RluF-RNA complex: a base-pair rearrangement is the key to selectivity of RluF for U2604 of the ribosome.J Mol Biol. 2009 May 15;388(4):785-800. doi: 10.1016/j.jmb.2009.03.029. Epub 2009 Mar 17. J Mol Biol. 2009. PMID: 19298824 Free PMC article.
-
Domain organization and crystal structure of the catalytic domain of E.coli RluF, a pseudouridine synthase that acts on 23S rRNA.J Mol Biol. 2006 Jun 16;359(4):998-1009. doi: 10.1016/j.jmb.2006.04.019. Epub 2006 Apr 25. J Mol Biol. 2006. PMID: 16712869
-
Crystal structure of the RluD pseudouridine synthase catalytic module, an enzyme that modifies 23S rRNA and is essential for normal cell growth of Escherichia coli.J Mol Biol. 2004 Jan 2;335(1):87-101. doi: 10.1016/j.jmb.2003.10.003. J Mol Biol. 2004. PMID: 14659742
-
The roles of the essential Asp-48 and highly conserved His-43 elucidated by the pH dependence of the pseudouridine synthase TruB.Arch Biochem Biophys. 2005 Jan 1;433(1):322-34. doi: 10.1016/j.abb.2004.09.009. Arch Biochem Biophys. 2005. PMID: 15581587 Review.
-
Substrate recognition by RNA 5-methyluridine methyltransferases and pseudouridine synthases: a structural perspective.J Biol Chem. 2006 Dec 22;281(51):38969-73. doi: 10.1074/jbc.R600034200. Epub 2006 Nov 3. J Biol Chem. 2006. PMID: 17085441 Review. No abstract available.
Cited by
-
The structural basis of mRNA recognition and binding by yeast pseudouridine synthase PUS1.PLoS One. 2023 Nov 8;18(11):e0291267. doi: 10.1371/journal.pone.0291267. eCollection 2023. PLoS One. 2023. PMID: 37939088 Free PMC article.
-
Accurate placement of substrate RNA by Gar1 in H/ACA RNA-guided pseudouridylation.Nucleic Acids Res. 2015 Sep 3;43(15):7207-16. doi: 10.1093/nar/gkv757. Epub 2015 Jul 22. Nucleic Acids Res. 2015. PMID: 26206671 Free PMC article.
-
The molecular basis of tRNA selectivity by human pseudouridine synthase 3.Mol Cell. 2024 Jul 11;84(13):2472-2489.e8. doi: 10.1016/j.molcel.2024.06.013. Mol Cell. 2024. PMID: 38996458 Free PMC article.
-
Pseudouridine in the Anticodon of Escherichia coli tRNATyr(QΨA) Is Catalyzed by the Dual Specificity Enzyme RluF.J Biol Chem. 2016 Oct 14;291(42):22327-22337. doi: 10.1074/jbc.M116.747865. Epub 2016 Aug 22. J Biol Chem. 2016. PMID: 27551044 Free PMC article.
-
Dye label interference with RNA modification reveals 5-fluorouridine as non-covalent inhibitor.Nucleic Acids Res. 2014 Nov 10;42(20):12735-45. doi: 10.1093/nar/gku908. Epub 2014 Oct 9. Nucleic Acids Res. 2014. PMID: 25300485 Free PMC article.
References
-
- Bjork G. Stable RNA Modification. In: Neidhardt FC, Curtiss R, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd edn. Washington, DC: ASM Press; 1996. pp. 861–886.
-
- Moore PB, Steitz TA. The involvement of RNA in ribosome function. Nature. 2002;418:229–235. - PubMed
-
- Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 2002;514:17–25. - PubMed
-
- Huang L, Pookanjanatavip M, Gu X, Santi DV. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry. 1998;37:344–351. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

