Negatome 2.0: a database of non-interacting proteins derived by literature mining, manual annotation and protein structure analysis
- PMID: 24214996
- PMCID: PMC3965096
- DOI: 10.1093/nar/gkt1079
Negatome 2.0: a database of non-interacting proteins derived by literature mining, manual annotation and protein structure analysis
Abstract
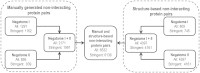
Knowledge about non-interacting proteins (NIPs) is important for training the algorithms to predict protein-protein interactions (PPIs) and for assessing the false positive rates of PPI detection efforts. We present the second version of Negatome, a database of proteins and protein domains that are unlikely to engage in physical interactions (available online at http://mips.helmholtz-muenchen.de/proj/ppi/negatome). Negatome is derived by manual curation of literature and by analyzing three-dimensional structures of protein complexes. The main methodological innovation in Negatome 2.0 is the utilization of an advanced text mining procedure to guide the manual annotation process. Potential non-interactions were identified by a modified version of Excerbt, a text mining tool based on semantic sentence analysis. Manual verification shows that nearly a half of the text mining results with the highest confidence values correspond to NIP pairs. Compared to the first version the contents of the database have grown by over 300%.
Figures
References
-
- Lees JG, Heriche JK, Morilla I, Ranea JA, Orengo CA. Systematic computational prediction of protein interaction networks. Phys. Biol. 2011;8:035008. - PubMed
-
- Trabuco LG, Betts MJ, Russell RB. Negative protein-protein interaction datasets derived from large-scale two-hybrid experiments. Methods. 2012;58:343–348. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

