DataHigh: graphical user interface for visualizing and interacting with high-dimensional neural activity
- PMID: 24216250
- PMCID: PMC3950756
- DOI: 10.1088/1741-2560/10/6/066012
DataHigh: graphical user interface for visualizing and interacting with high-dimensional neural activity
Abstract
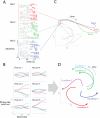
Objective: Analyzing and interpreting the activity of a heterogeneous population of neurons can be challenging, especially as the number of neurons, experimental trials, and experimental conditions increases. One approach is to extract a set of latent variables that succinctly captures the prominent co-fluctuation patterns across the neural population. A key problem is that the number of latent variables needed to adequately describe the population activity is often greater than 3, thereby preventing direct visualization of the latent space. By visualizing a small number of 2-d projections of the latent space or each latent variable individually, it is easy to miss salient features of the population activity.
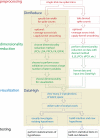
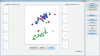
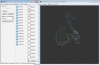
Approach: To address this limitation, we developed a Matlab graphical user interface (called DataHigh) that allows the user to quickly and smoothly navigate through a continuum of different 2-d projections of the latent space. We also implemented a suite of additional visualization tools (including playing out population activity timecourses as a movie and displaying summary statistics, such as covariance ellipses and average timecourses) and an optional tool for performing dimensionality reduction.
Main results: To demonstrate the utility and versatility of DataHigh, we used it to analyze single-trial spike count and single-trial timecourse population activity recorded using a multi-electrode array, as well as trial-averaged population activity recorded using single electrodes.
Significance: DataHigh was developed to fulfil a need for visualization in exploratory neural data analysis, which can provide intuition that is critical for building scientific hypotheses and models of population activity.
Figures










References
-
- Asimov D. The grand tour: a tool for viewing multidimensional data. SIAM Journal on Scientific and Statistical Computing. 1985;6(1):128–143.
-
- Averbeck BB, Latham PE, Pouget A. Neural correlations, population coding and computation. Nature Reviews Neuroscience. 2006;7(5):358–366. - PubMed
-
- Berk R, Brown L, Zhao L. Statistical inference after model selection. Journal of Quantitative Criminology. 2010;26(2):217–236.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
