Allogeneic T cell responses are regulated by a specific miRNA-mRNA network
- PMID: 24216511
- PMCID: PMC3809794
- DOI: 10.1172/JCI70013
Allogeneic T cell responses are regulated by a specific miRNA-mRNA network
Abstract
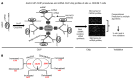
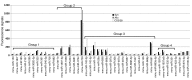
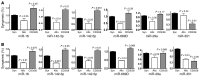
Donor T cells that respond to host alloantigens following allogeneic bone marrow transplantation (BMT) induce graft-versus-host (GVH) responses, but their molecular landscape is not well understood. MicroRNAs (miRNAs) regulate gene (mRNA) expression and fine-tune the molecular responses of T cells. We stimulated naive T cells with either allogeneic or nonspecific stimuli and used argonaute cross-linked immunoprecipitation (CLIP) with subsequent ChIP microarray analyses to profile miR responses and their direct mRNA targets. We identified a unique expression pattern of miRs and mRNAs following the allostimulation of T cells and a high correlation between the expression of the identified miRs and a reduction of their mRNA targets. miRs and mRNAs that were predicted to be differentially regulated in allogeneic T cells compared with nonspecifically stimulated T cells were validated in vitro. These analyses identified wings apart-like homolog (Wapal) and synaptojanin 1 (Synj1) as potential regulators of allogeneic T cell responses. The expression of these molecular targets in vivo was confirmed in MHC-mismatched experimental BMT. Targeted silencing of either Wapal or Synj1 prevented the development of GVH response, confirming a role for these regulators in allogeneic T cell responses. Thus, this genome-wide analysis of miRNA-mRNA interactions identifies previously unrecognized molecular regulators of T cell responses.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

