Epigenetics and colorectal cancer pathogenesis
- PMID: 24216997
- PMCID: PMC3730326
- DOI: 10.3390/cancers5020676
Epigenetics and colorectal cancer pathogenesis
Abstract
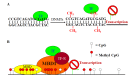
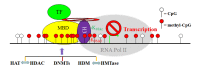
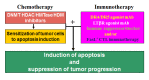
Colorectal cancer (CRC) develops through a multistage process that results from the progressive accumulation of genetic mutations, and frequently as a result of mutations in the Wnt signaling pathway. However, it has become evident over the past two decades that epigenetic alterations of the chromatin, particularly the chromatin components in the promoter regions of tumor suppressors and oncogenes, play key roles in CRC pathogenesis. Epigenetic regulation is organized at multiple levels, involving primarily DNA methylation and selective histone modifications in cancer cells. Assessment of the CRC epigenome has revealed that virtually all CRCs have aberrantly methylated genes and that the average CRC methylome has thousands of abnormally methylated genes. Although relatively less is known about the patterns of specific histone modifications in CRC, selective histone modifications and resultant chromatin conformation have been shown to act, in concert with DNA methylation, to regulate gene expression to mediate CRC pathogenesis. Moreover, it is now clear that not only DNA methylation but also histone modifications are reversible processes. The increased understanding of epigenetic regulation of gene expression in the context of CRC pathogenesis has led to development of epigenetic biomarkers for CRC diagnosis and epigenetic drugs for CRC therapy.
Figures
References
-
- Kopetz S., Chang G.J., Overman M.J., Eng C., Sargent D.J., Larson D.W., Grothey A., Vauthey J.N., Nagorney D.M., McWilliams R.R. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J. Clin. Oncol. 2009;27:3677–3683. doi: 10.1200/JCO.2008.20.5278. - DOI - PMC - PubMed
-
- Terzic J., Grivennikov S., Karin E., Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114.e5. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

