Genetic factors involved in risk for methamphetamine intake and sensitization
- PMID: 24217691
- PMCID: PMC3880562
- DOI: 10.1007/s00335-013-9484-9
Genetic factors involved in risk for methamphetamine intake and sensitization
Abstract
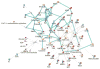
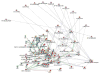
Lines of mice were created by selective breeding for the purpose of identifying genetic mechanisms that influence the magnitude of the selected trait and to explore genetic correlations for additional traits thought to be influenced by shared mechanisms. DNA samples from high and low methamphetamine-drinking (MADR) and high and low methamphetamine-sensitization lines were used for quantitative trait locus (QTL) mapping. Significant additive genetic correlations between the two traits indicated a common genetic influence, and a QTL on chromosome X was detected for both traits, suggesting one source of this commonality. For MADR mice, a QTL on chromosome 10 accounted for more than 50 % of the genetic variance in that trait. Microarray gene expression analyses were performed for three brain regions for methamphetamine-naïve MADR line mice: nucleus accumbens, prefrontal cortex, and ventral midbrain. Many of the genes that were differentially expressed between the high and low MADR lines were shared in common across the three brain regions. A gene network highly enriched in transcription factor genes was identified as being relevant to genetically determined differences in methamphetamine intake. When the mu opioid receptor gene (Oprm1), located on chromosome 10 in the QTL region, was added to this top-ranked transcription factor network, it became a hub in the network. These data are consistent with previously published findings of opioid response and intake differences between the MADR lines and suggest that Oprm1, or a gene that impacts activity of the opioid system, plays a role in genetically determined differences in methamphetamine intake.
Figures




References
-
- Belknap JK, Hitzemann R, Crabbe JC, Phillips TJ, Buck KJ, Williams RW. QTL analysis and genomewide mutagenesis in mice: complementary genetic approaches to the dissection of complex traits. Behav Genet. 2001;31:5–15. - PubMed
-
- Belknap JK, Mitchell SR, O’Toole LA, Helms ML, Crabbe JC. Type I and type II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains. Behav Genet. 1996;26:149–160. - PubMed
-
- Belknap JK, Richards SP, O’Toole LA, Helms ML, Phillips TJ. Short-term selective breeding as a tool for QTL mapping: Ethanol preference drinking in mice. Behav Genet. 1997;27:55–66. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

