The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data
- PMID: 24217912
- PMCID: PMC3965098
- DOI: 10.1093/nar/gkt1026
The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data
Abstract
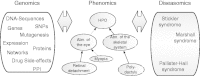
The Human Phenotype Ontology (HPO) project, available at http://www.human-phenotype-ontology.org, provides a structured, comprehensive and well-defined set of 10,088 classes (terms) describing human phenotypic abnormalities and 13,326 subclass relations between the HPO classes. In addition we have developed logical definitions for 46% of all HPO classes using terms from ontologies for anatomy, cell types, function, embryology, pathology and other domains. This allows interoperability with several resources, especially those containing phenotype information on model organisms such as mouse and zebrafish. Here we describe the updated HPO database, which provides annotations of 7,278 human hereditary syndromes listed in OMIM, Orphanet and DECIPHER to classes of the HPO. Various meta-attributes such as frequency, references and negations are associated with each annotation. Several large-scale projects worldwide utilize the HPO for describing phenotype information in their datasets. We have therefore generated equivalence mappings to other phenotype vocabularies such as LDDB, Orphanet, MedDRA, UMLS and phenoDB, allowing integration of existing datasets and interoperability with multiple biomedical resources. We have created various ways to access the HPO database content using flat files, a MySQL database, and Web-based tools. All data and documentation on the HPO project can be found online.
Figures

References
-
- Amberger J, Bocchini CA, Hamosh A. A new face and new challenges for online mendelian inheritance in man (OMIM®) Hum. Mutat. 2011;32:564–567. - PubMed
-
- Doelken SC, Köhler S, Mungall CJ, Gkoutos GV, Ruef BJ, Smith C, Smedley D, Bauer S, Klopocki E, Schofield PN, et al. Phenotypic overlap in the contribution of individual genes to CNV pathogenicity revealed by cross-species computational analysis of single-gene mutations in humans, mice and zebrafish. Dis. Model Mech. 2013;6:358–372. - PMC - PubMed
-
- Biesecker LG. Phenotype matters. Nat. Genet. 2004;36:323–324. - PubMed
-
- Robinson PN. Deep phenotyping for precision medicine. Hum. Mutat. 2012;33:777–780. - PubMed
Publication types
MeSH terms
Grants and funding
- RP-PG-0310-1002/DH_/Department of Health/United Kingdom
- R24-OD011883/OD/NIH HHS/United States
- R01-HG004838/HG/NHGRI NIH HHS/United States
- U41 HG000330/HG/NHGRI NIH HHS/United States
- R24 OD011883/OD/NIH HHS/United States
- U41 HG002659/HG/NHGRI NIH HHS/United States
- RG/09/012/28096/BHF_/British Heart Foundation/United Kingdom
- MC_PC_U127580972/MRC_/Medical Research Council/United Kingdom
- HG000330/HG/NHGRI NIH HHS/United States
- R01 MH074090/MH/NIMH NIH HHS/United States
- P41 HG002659/HG/NHGRI NIH HHS/United States
- U41-HG002659/HG/NHGRI NIH HHS/United States
- MC_PC_U127561093/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

