Finding the target sites of RNA-binding proteins
- PMID: 24217996
- PMCID: PMC4253089
- DOI: 10.1002/wrna.1201
Finding the target sites of RNA-binding proteins
Abstract
RNA-protein interactions differ from DNA-protein interactions because of the central role of RNA secondary structure. Some RNA-binding domains (RBDs) recognize their target sites mainly by their shape and geometry and others are sequence-specific but are sensitive to secondary structure context. A number of small- and large-scale experimental approaches have been developed to measure RNAs associated in vitro and in vivo with RNA-binding proteins (RBPs). Generalizing outside of the experimental conditions tested by these assays requires computational motif finding. Often RBP motif finding is done by adapting DNA motif finding methods; but modeling secondary structure context leads to better recovery of RBP-binding preferences. Genome-wide assessment of mRNA secondary structure has recently become possible, but these data must be combined with computational predictions of secondary structure before they add value in predicting in vivo binding. There are two main approaches to incorporating structural information into motif models: supplementing primary sequence motif models with preferred secondary structure contexts (e.g., MEMERIS and RNAcontext) and directly modeling secondary structure recognized by the RBP using stochastic context-free grammars (e.g., CMfinder and RNApromo). The former better reconstruct known binding preferences for sequence-specific RBPs but are not suitable for modeling RBPs that recognize shape and geometry of RNAs. Future work in RBP motif finding should incorporate interactions between multiple RBDs and multiple RBPs in binding to RNA.
© 2013 John Wiley & Sons, Ltd.
Figures
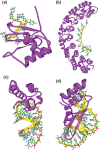
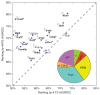
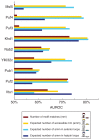
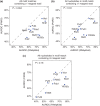

References
-
- Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. - PubMed
-
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. - PubMed
-
- Seeman NC, Rosenberg JM, Suddath FL, Kim JJ, Rich A. RNA double-helical fragments at atomic resolution. I. The crystal and molecular structure of sodium adenylyl-3′,5′-uridine hexahydrate. J Mol Biol. 1976;104:109–144. - PubMed
FURTHER READING
-
- Messias AC, Sattler M. Structural basis of single-stranded RNA recognition. Acc Chem Res. 2004;37:279–287. - PubMed
-
- Chang KY, Ramos A. The double-stranded RNA-binding motif a versatile macromolecular docking platform. FEBS J. 2005;272:2109–2117. - PubMed
-
- Fierro-Monti I, Mathews MB. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem Sci. 2000;25:241–246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

