The distinctive germinal center phase of IgE+ B lymphocytes limits their contribution to the classical memory response
- PMID: 24218137
- PMCID: PMC3832920
- DOI: 10.1084/jem.20131539
The distinctive germinal center phase of IgE+ B lymphocytes limits their contribution to the classical memory response
Abstract
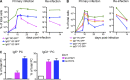
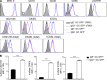
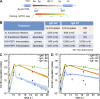
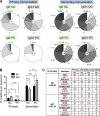
The mechanisms involved in the maintenance of memory IgE responses are poorly understood, and the role played by germinal center (GC) IgE(+) cells in memory responses is particularly unclear. IgE(+) B cell differentiation is characterized by a transient GC phase, a bias toward the plasma cell (PC) fate, and dependence on sequential switching for the production of high-affinity IgE. We show here that IgE(+) GC B cells are unfit to undergo the conventional GC differentiation program due to impaired B cell receptor function and increased apoptosis. IgE(+) GC cells fail to populate the GC light zone and are unable to contribute to the memory and long-lived PC compartments. Furthermore, we demonstrate that direct and sequential switching are linked to distinct B cell differentiation fates: direct switching generates IgE(+) GC cells, whereas sequential switching gives rise to IgE(+) PCs. We propose a comprehensive model for the generation and memory of IgE responses.
Figures






References
-
- Camberis M., Le Gros G., Urban J., Jr 2003. Animal model of Nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr. Protoc. Immunol. Chapter 19:12. - PubMed
-
- Curotto de Lafaille M.A., Lafaille J.J. 2010. The biology of IgE: The generation of High Affinity IgE Antibodies. Cancer and IgE: Introducing the Concept of AllergoOncology. Penichet M.L., Jensen-Jarolim E., Springer; 37–46
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

