A collagen-remodeling gene signature regulated by TGF-β signaling is associated with metastasis and poor survival in serous ovarian cancer
- PMID: 24218511
- PMCID: PMC3946428
- DOI: 10.1158/1078-0432.CCR-13-1256
A collagen-remodeling gene signature regulated by TGF-β signaling is associated with metastasis and poor survival in serous ovarian cancer
Abstract
Purpose: To elucidate molecular pathways contributing to metastatic cancer progression and poor clinical outcome in serous ovarian cancer.
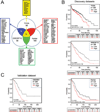
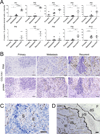
Experimental design: Poor survival signatures from three different serous ovarian cancer datasets were compared and a common set of genes was identified. The predictive value of this gene signature was validated in independent datasets. The expression of the signature genes was evaluated in primary, metastatic, and/or recurrent cancers using quantitative PCR and in situ hybridization. Alterations in gene expression by TGF-β1 and functional consequences of loss of COL11A1 were evaluated using pharmacologic and knockdown approaches, respectively.
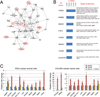
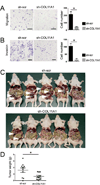
Results: We identified and validated a 10-gene signature (AEBP1, COL11A1, COL5A1, COL6A2, LOX, POSTN, SNAI2, THBS2, TIMP3, and VCAN) that is associated with poor overall survival (OS) in patients with high-grade serous ovarian cancer. The signature genes encode extracellular matrix proteins involved in collagen remodeling. Expression of the signature genes is regulated by TGF-β1 signaling and is enriched in metastases in comparison with primary ovarian tumors. We demonstrate that levels of COL11A1, one of the signature genes, continuously increase during ovarian cancer disease progression, with the highest expression in recurrent metastases. Knockdown of COL11A1 decreases in vitro cell migration, invasion, and tumor progression in mice.
Conclusion: Our findings suggest that collagen-remodeling genes regulated by TGF-β1 signaling promote metastasis and contribute to poor OS in patients with serous ovarian cancer. Our 10-gene signature has both predictive value and biologic relevance and thus may be useful as a therapeutic target.
©2013 AACR.
Conflict of interest statement
Figures

References
-
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. - PubMed
-
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. - PubMed
-
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

