Many human accelerated regions are developmental enhancers
- PMID: 24218637
- PMCID: PMC3826498
- DOI: 10.1098/rstb.2013.0025
Many human accelerated regions are developmental enhancers
Abstract
The genetic changes underlying the dramatic differences in form and function between humans and other primates are largely unknown, although it is clear that gene regulatory changes play an important role. To identify regulatory sequences with potentially human-specific functions, we and others used comparative genomics to find non-coding regions conserved across mammals that have acquired many sequence changes in humans since divergence from chimpanzees. These regions are good candidates for performing human-specific regulatory functions. Here, we analysed the DNA sequence, evolutionary history, histone modifications, chromatin state and transcription factor (TF) binding sites of a combined set of 2649 non-coding human accelerated regions (ncHARs) and predicted that at least 30% of them function as developmental enhancers. We prioritized the predicted ncHAR enhancers using analysis of TF binding site gain and loss, along with the functional annotations and expression patterns of nearby genes. We then tested both the human and chimpanzee sequence for 29 ncHARs in transgenic mice, and found 24 novel developmental enhancers active in both species, 17 of which had very consistent patterns of activity in specific embryonic tissues. Of these ncHAR enhancers, five drove expression patterns suggestive of different activity for the human and chimpanzee sequence at embryonic day 11.5. The changes to human non-coding DNA in these ncHAR enhancers may modify the complex patterns of gene expression necessary for proper development in a human-specific manner and are thus promising candidates for understanding the genetic basis of human-specific biology.
Keywords: development; enhancers; gene regulation; human accelerated regions; primate evolution.
Figures


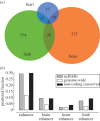

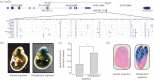

References
-
- Chimpanzee Sequencing and Analysis Consortium 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 (doi:10.1038/nature04072) - DOI - PubMed
-
- Jiang Z, Tang H, Ventura M, Cardone M, Marques-Bonet T, She X, Pevzner P, Eichler E. 2007. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 39, 1361–1368 (doi:10.1038/ng.2007.9) - DOI - PubMed
-
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (doi:10.1101/gr.097857.109) - DOI - PMC - PubMed
-
- Lindblad-Toh K, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (doi:10.1038/nature10530) - DOI - PMC - PubMed
-
- Church DM, et al. 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 7, e1000112 (doi:10.1371/journal.pbio.1000112) - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

