Conditional Dicer substrate formation via shape and sequence transduction with small conditional RNAs
- PMID: 24219616
- PMCID: PMC3842090
- DOI: 10.1021/ja404676x
Conditional Dicer substrate formation via shape and sequence transduction with small conditional RNAs
Abstract
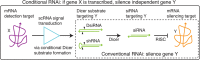
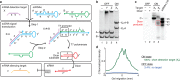
RNA interference (RNAi) mediated by small interfering RNAs (siRNAs) enables knockdown of a gene of choice, executing the logical operation: silence gene Y. The fact that the siRNA is constitutively active is a significant limitation, making it difficult to confine knockdown to a specific locus and time. To achieve spatiotemporal control over silencing, we seek to engineer small conditional RNAs (scRNAs) that mediate 'conditional RNAi' corresponding to the logical operation: if gene X is transcribed, silence independent gene Y. By appropriately selecting gene X, knockdown of gene Y could then be restricted in a tissue- and time-specific manner. To implement the logic of conditional RNAi, our approach is to engineer scRNAs that, upon binding to mRNA 'detection target' X, perform shape and sequence transduction to form a Dicer substrate targeting independent mRNA 'silencing target' Y, with subsequent Dicer processing yielding an siRNA targeting mRNA Y for destruction. Toward this end, here we design and experimentally validate diverse scRNA mechanisms for conditional Dicer substrate formation. Test tube studies demonstrate strong OFF/ON conditional response, with at least an order of magnitude increase in Dicer substrate production in the presence of the cognate mRNA detection target. By appropriately dimensioning and/or chemically modifying the scRNAs, only the product of signal transduction, and not the reactants or intermediates, is efficiently processed by Dicer, yielding siRNAs. These mechanism studies explore diverse design principles for engineering scRNA signal transduction cascades including reactant stability vs metastability, catalytic vs noncatalytic transduction, pre- vs post-transcriptional transduction, reactant and product molecularity, and modes of molecular self-assembly and disassembly.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

