DNA-dependent protein kinase regulates DNA end resection in concert with Mre11-Rad50-Nbs1 (MRN) and ataxia telangiectasia-mutated (ATM)
- PMID: 24220101
- PMCID: PMC3873567
- DOI: 10.1074/jbc.M113.514398
DNA-dependent protein kinase regulates DNA end resection in concert with Mre11-Rad50-Nbs1 (MRN) and ataxia telangiectasia-mutated (ATM)
Abstract
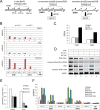
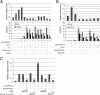
The resection of DNA double strand breaks initiates homologous recombination (HR) and is critical for genomic stability. Using direct measurement of resection in human cells and reconstituted assays of resection with purified proteins in vitro, we show that DNA-dependent protein kinase catalytic subunit (DNA-PKcs), a classic nonhomologous end joining factor, antagonizes double strand break resection by blocking the recruitment of resection enzymes such as exonuclease 1 (Exo1). Autophosphorylation of DNA-PKcs promotes DNA-PKcs dissociation and consequently Exo1 binding. Ataxia telangiectasia-mutated kinase activity can compensate for DNA-PKcs autophosphorylation and promote resection under conditions where DNA-PKcs catalytic activity is inhibited. The Mre11-Rad50-Nbs1 (MRN) complex further stimulates resection in the presence of Ku and DNA-PKcs by recruiting Exo1 and enhancing DNA-PKcs autophosphorylation, and it also inhibits DNA ligase IV/XRCC4-mediated end rejoining. This work suggests that, in addition to its key role in nonhomologous end joining, DNA-PKcs also acts in concert with MRN and ataxia telangiectasia-mutated to regulate resection and thus DNA repair pathway choice.
Keywords: DNA Damage Response; DNA Enzymes; DNA Recombination; DNA Repair; DNA-binding Protein; Protein Kinases.
Figures





Similar articles
-
Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex.J Biol Chem. 2013 May 3;288(18):12840-51. doi: 10.1074/jbc.M113.460378. Epub 2013 Mar 22. J Biol Chem. 2013. PMID: 23525106 Free PMC article.
-
Localization of Double-Strand Break Repair Proteins to Viral Replication Compartments following Lytic Reactivation of Kaposi's Sarcoma-Associated Herpesvirus.J Virol. 2017 Oct 27;91(22):e00930-17. doi: 10.1128/JVI.00930-17. Print 2017 Nov 15. J Virol. 2017. PMID: 28855246 Free PMC article.
-
Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of nonhomologous end joining (NHEJ) in G1 and resection-mediated inhibition of NHEJ in G2.Mol Cell Biol. 2009 Oct;29(20):5552-63. doi: 10.1128/MCB.00476-09. Epub 2009 Aug 10. Mol Cell Biol. 2009. PMID: 19667071 Free PMC article.
-
Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template.Biochem Cell Biol. 2007 Aug;85(4):509-20. doi: 10.1139/O07-069. Biochem Cell Biol. 2007. PMID: 17713585 Review.
-
MRN complex is an essential effector of DNA damage repair.J Zhejiang Univ Sci B. 2021 Jan 15;22(1):31-37. doi: 10.1631/jzus.B2000289. J Zhejiang Univ Sci B. 2021. PMID: 33448185 Free PMC article. Review.
Cited by
-
Intrinsic ATR signaling shapes DNA end resection and suppresses toxic DNA-PKcs signaling.NAR Cancer. 2020 Jun;2(2):zcaa006. doi: 10.1093/narcan/zcaa006. Epub 2020 May 1. NAR Cancer. 2020. PMID: 32743550 Free PMC article.
-
SV40 utilizes ATM kinase activity to prevent non-homologous end joining of broken viral DNA replication products.PLoS Pathog. 2014 Dec 4;10(12):e1004536. doi: 10.1371/journal.ppat.1004536. eCollection 2014 Dec. PLoS Pathog. 2014. PMID: 25474690 Free PMC article.
-
Non-redundant Functions of ATM and DNA-PKcs in Response to DNA Double-Strand Breaks.Cell Rep. 2015 Nov 24;13(8):1598-609. doi: 10.1016/j.celrep.2015.10.024. Epub 2015 Nov 12. Cell Rep. 2015. PMID: 26586426 Free PMC article.
-
Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.Mol Cell. 2022 Jan 6;82(1):177-189.e4. doi: 10.1016/j.molcel.2021.11.025. Epub 2021 Dec 21. Mol Cell. 2022. PMID: 34936881 Free PMC article.
-
TIP60 K430 SUMOylation attenuates its interaction with DNA-PKcs in S-phase cells: Facilitating homologous recombination and emerging target for cancer therapy.Sci Adv. 2020 Jul 10;6(28):eaba7822. doi: 10.1126/sciadv.aba7822. eCollection 2020 Jul. Sci Adv. 2020. PMID: 32832608 Free PMC article.
References
-
- Trovesi C., Manfrini N., Falcettoni M., Longhese M. P. (2013) Regulation of the DNA damage response by cyclin-dependent kinases. J. Mol. Biol. 425, 4756–4766 - PubMed
-
- Symington L. S., Gautier J. (2011) Double strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

