Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors
- PMID: 24220501
- PMCID: PMC4389866
- DOI: 10.4161/auto.26841
Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors
Abstract
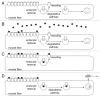
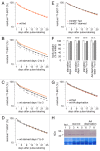
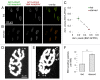
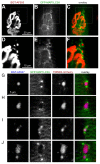
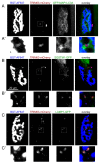
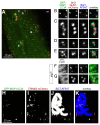
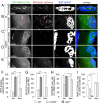
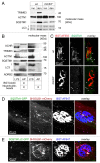
Removal of ubiquitinated targets by autophagosomes can be mediated by receptor molecules, like SQSTM1, in a mechanism referred to as selective autophagy. While cytoplasmic protein aggregates, mitochondria, and bacteria are the best-known targets of selective autophagy, their role in the turnover of membrane receptors is scarce. We here showed that fasting-induced wasting of skeletal muscle involves remodeling of the neuromuscular junction (NMJ) by increasing the turnover of muscle-type CHRN (cholinergic receptor, nicotinic/nicotinic acetylcholine receptor) in a TRIM63-dependent manner. Notably, this process implied enhanced production of endo/lysosomal carriers of CHRN, which also contained the membrane remodeler SH3GLB1, the E3 ubiquitin ligase, TRIM63, and the selective autophagy receptor SQSTM1. Furthermore, these vesicles were surrounded by the autophagic marker MAP1LC3A in an ATG7-dependent fashion, and some of them were also positive for the lysosomal marker, LAMP1. While the amount of vesicles containing endocytosed CHRN strongly augmented in the absence of ATG7 as well as upon denervation as a model for long-term atrophy, denervation-induced increase in autophagic CHRN vesicles was completely blunted in the absence of TRIM63. On a similar note, in trim63(-/-) mice denervation-induced upregulation of SQSTM1 and LC3-II was abolished and endogenous SQSTM1 did not colocalize with CHRN vesicles as it did in the wild type. SQSTM1 and LC3-II coprecipitated with surface-labeled/endocytosed CHRN and SQSTM1 overexpression significantly induced CHRN vesicle formation. Taken together, our data suggested that selective autophagy regulates the basal and atrophy-induced turnover of the pentameric transmembrane protein, CHRN, and that TRIM63, together with SH3GLB1 and SQSTM1 regulate this process.
Keywords: Bif-1; LC3; MuRF1; SQSTM1/p62; acetylcholine receptor; atrophy; endophilin B1; neuromuscular junction; selective autophagy.
Figures
Similar articles
-
Progress of endocytic CHRN to autophagic degradation is regulated by RAB5-GTPase and T145 phosphorylation of SH3GLB1 at mouse neuromuscular junctions in vivo.Autophagy. 2016 Dec;12(12):2300-2310. doi: 10.1080/15548627.2016.1234564. Epub 2016 Oct 7. Autophagy. 2016. PMID: 27715385 Free PMC article.
-
Regulation of nicotinic acetylcholine receptor turnover by MuRF1 connects muscle activity to endo/lysosomal and atrophy pathways.Age (Dordr). 2013 Oct;35(5):1663-74. doi: 10.1007/s11357-012-9468-9. Epub 2012 Sep 6. Age (Dordr). 2013. PMID: 22956146 Free PMC article.
-
Developmental regulation of MURF ubiquitin ligases and autophagy proteins nbr1, p62/SQSTM1 and LC3 during cardiac myofibril assembly and turnover.Dev Biol. 2011 Mar 1;351(1):46-61. doi: 10.1016/j.ydbio.2010.12.024. Epub 2010 Dec 23. Dev Biol. 2011. PMID: 21185285 Free PMC article.
-
Regulation of selective autophagy: the p62/SQSTM1 paradigm.Essays Biochem. 2017 Dec 12;61(6):609-624. doi: 10.1042/EBC20170035. Print 2017 Dec 12. Essays Biochem. 2017. PMID: 29233872 Review.
-
The Metabolic Stability of the Nicotinic Acetylcholine Receptor at the Neuromuscular Junction.Cells. 2021 Feb 9;10(2):358. doi: 10.3390/cells10020358. Cells. 2021. PMID: 33572348 Free PMC article. Review.
Cited by
-
The TOR Pathway at the Neuromuscular Junction: More Than a Metabolic Player?Front Mol Neurosci. 2020 Aug 28;13:162. doi: 10.3389/fnmol.2020.00162. eCollection 2020. Front Mol Neurosci. 2020. PMID: 32982690 Free PMC article. Review.
-
Do neurogenic and cancer-induced muscle atrophy follow common or divergent paths?Eur J Transl Myol. 2018 Dec 13;28(4):7931. doi: 10.4081/ejtm.2018.7931. eCollection 2018 Nov 2. Eur J Transl Myol. 2018. PMID: 30662704 Free PMC article.
-
Variations of chromosomes 2 and 3 gene expression profiles among pulmonary telocytes, pneumocytes, airway cells, mesenchymal stem cells and lymphocytes.J Cell Mol Med. 2014 Oct;18(10):2044-60. doi: 10.1111/jcmm.12429. Epub 2014 Oct 2. J Cell Mol Med. 2014. PMID: 25278030 Free PMC article.
-
Neuromuscular junction degeneration in muscle wasting.Curr Opin Clin Nutr Metab Care. 2016 May;19(3):177-81. doi: 10.1097/MCO.0000000000000267. Curr Opin Clin Nutr Metab Care. 2016. PMID: 26870889 Free PMC article. Review.
-
Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1.Autophagy. 2021 Jan;17(1):1-382. doi: 10.1080/15548627.2020.1797280. Epub 2021 Feb 8. Autophagy. 2021. PMID: 33634751 Free PMC article.
References
-
- Shaid S, Brandts CH, Serve H, Dikic I. . Ubiquitination and selective autophagy. Cell Death Differ 2013; 20:21 - 30; http://dx.doi.org/10.1038/cdd.2012.72; PMID: 22722335 - DOI - PMC - PubMed
-
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. . p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171:603 - 14; http://dx.doi.org/10.1083/jcb.200507002; PMID: 16286508 - DOI - PMC - PubMed
-
- Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. . . A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009; 33:505 - 16; http://dx.doi.org/10.1016/j.molcel.2009.01.020; PMID: 19250911 - DOI - PubMed
-
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. . p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131 - 45; http://dx.doi.org/10.1074/jbc.M702824200; PMID: 17580304 - DOI - PubMed
-
- Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K. . Alteration of autophagosomal proteins (LC3, GABARAP and GATE-16) in Lewy body disease. Neurobiol Dis 2011; 43:690 - 7; http://dx.doi.org/10.1016/j.nbd.2011.05.022; PMID: 21684337 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
