Transcriptional control of hepatic lipid metabolism by SREBP and ChREBP
- PMID: 24222088
- PMCID: PMC4035704
- DOI: 10.1055/s-0033-1358523
Transcriptional control of hepatic lipid metabolism by SREBP and ChREBP
Abstract
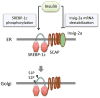
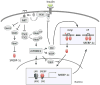
The liver is a central organ that controls systemic energy homeostasis and nutrient metabolism. Dietary carbohydrates and lipids, and fatty acids derived from adipose tissue are delivered to the liver, and utilized for gluconeogenesis, lipogenesis, and ketogenesis, which are tightly regulated by hormonal and neural signals. Hepatic lipogenesis is activated primarily by insulin that is secreted from the pancreas after a high-carbohydrate meal. Sterol regulatory element binding protein-1c (SREBP-1c) and carbohydrate-responsive element-binding protein (ChREBP) are major transcriptional regulators that induce key lipogenic enzymes to promote lipogenesis in the liver. Sterol regulatory element binding protein-1c is activated by insulin through complex signaling cascades that control SREBP-1c at both transcriptional and posttranslational levels. Carbohydrate-responsive element-binding protein is activated by glucose independently of insulin. Here, the authors attempt to summarize the current understanding of the molecular mechanism for the transcriptional regulation of hepatic lipogenesis, focusing on recent studies that explore the signaling pathways controlling SREBPs and ChREBP.
Thieme Medical Publishers 333 Seventh Avenue, New York, NY 10001, USA.
Figures


References
-
- Towle HC. Glucose as a regulator of eukaryotic gene transcription. Trends in endocrinology and metabolism: TEM. 2005;16(10):489–494. - PubMed
-
- Liang G, Yang J, Horton JD, et al. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem. 2002;277(11):9520–9528. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

