Determining the binding affinity of therapeutic monoclonal antibodies towards their native unpurified antigens in human serum
- PMID: 24223227
- PMCID: PMC3819287
- DOI: 10.1371/journal.pone.0080501
Determining the binding affinity of therapeutic monoclonal antibodies towards their native unpurified antigens in human serum
Abstract
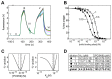
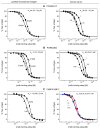
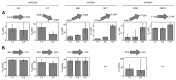
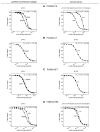
Monoclonal antibodies (mAbs) are a growing segment of therapeutics, yet their in vitro characterization remains challenging. While it is essential that a therapeutic mAb recognizes the native, physiologically occurring epitope, the generation and selection of mAbs often rely on the use of purified recombinant versions of the antigen that may display non-native epitopes. Here, we present a method to measure both, the binding affinity of a therapeutic mAb towards its native unpurified antigen in human serum, and the antigen's endogenous concentration, by combining the kinetic exclusion assay and Biacore's calibration free concentration analysis. To illustrate the broad utility of our method, we studied a panel of mAbs raised against three disparate soluble antigens that are abundant in the serum of healthy donors: proprotein convertase subtilisin/kexin type 9 (PCSK9), progranulin (PGRN), and fatty acid binding protein (FABP4). We also determined the affinity of each mAb towards its purified recombinant antigen and assessed whether the interactions were pH-dependent. Of the six mAbs studied, three did not appear to discriminate between the serum and recombinant forms of the antigen; one mAb bound serum antigen with a higher affinity than recombinant antigen; and two mAbs displayed a different affinity for serum antigen that could be explained by a pH-dependent interaction. Our results highlight the importance of taking pH into account when measuring the affinities of mAbs towards their serum antigens, since the pH of serum samples becomes increasingly alkaline upon aerobic handling.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

