Four Promoters of IRF5 Respond Distinctly to Stimuli and are Affected by Autoimmune-Risk Polymorphisms
- PMID: 24223576
- PMCID: PMC3819785
- DOI: 10.3389/fimmu.2013.00360
Four Promoters of IRF5 Respond Distinctly to Stimuli and are Affected by Autoimmune-Risk Polymorphisms
Abstract
Introduction: Autoimmune diseases such as systemic lupus erythematosus, rheumatoid arthritis, and multiple sclerosis affect millions of people worldwide. Interferon regulatory factor 5 (IRF5) contains polymorphisms associated with these autoimmune diseases. Two of these functional polymorphisms are found upstream of the IRF5 gene. rs2004640, which is a single nucleotide polymorphism and the CGGGG insertion/deletion (indel) were studied. IRF5 uses four different promoters for its four first exons: 1A, 1B, 1C, and 1D. Each promoter was analyzed, including functional differences due to the autoimmune-risk polymorphisms.
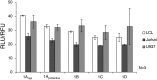
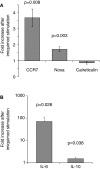
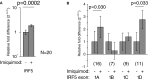
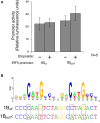
Results: IRF5 promoters were analyzed using ChIP-Seq data (ENCODE database) and the FactorBook database to define transcription factor binding sites. To verify promoter activity, the promoters were cloned into luciferase plasmids. Each construct exhibited luciferase activity. Exons 1A and 1D contain putative PU.1 and NFkB binding sites. Imiquimod, a Toll-like receptor 7 (TLR7) ligand, was used to activate these transcription factors. IRF5 levels were doubled after imiquimod treatment (p < 0.001), with specific increases in the 1A promoter (2.2-fold, p = 0.03) and 1D promoter (2.8-fold, p = 0.03). A putative binding site for p53, which affects apoptosis, was found in the promoter for exon 1B. However, site-directed mutagenesis of the p53 site showed no effect in a reporter assay.
Conclusion: The IRF5 exon 1B promoter has been characterized, and the responses of each IRF5 promoter to TLR7 stimulation have been determined. Changes in promoter activity and gene expression are likely due to specific and distinct transcription factors that bind to each promoter. Since high expression of IRF5 contributes to the development of autoimmune disease, understanding the source of increased IRF5 levels is key to understanding autoimmune etiology.
Keywords: IRF5; alternative promoters; autoimmune disease risk; interferon; systemic lupus erythematosus.
Figures



References
-
- Barnes BJ, Kellum MJ, Pinder KE, Frisancho JA, Pitha PM. Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res (2003) 63(19):6424–31 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

