Crystal structure of a four-layer aggregate of engineered TMV CP implies the importance of terminal residues for oligomer assembly
- PMID: 24223721
- PMCID: PMC3817195
- DOI: 10.1371/journal.pone.0077717
Crystal structure of a four-layer aggregate of engineered TMV CP implies the importance of terminal residues for oligomer assembly
Abstract
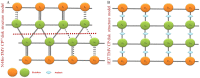
Background: Crystal structures of the tobacco mosaic virus (TMV) coat protein (CP) in its helical and disk conformations have previously been determined at the atomic level. For the helical structure, interactions of proteins and nucleic acids in the main chains were clearly observed; however, the conformation of residues at the C-terminus was flexible and disordered. For the four-layer aggregate disk structure, interactions of the main chain residues could only be observed through water-mediated hydrogen bonding with protein residues. In this study, the effects of the C-terminal peptides on the interactions of TMV CP were investigated by crystal structure determination.
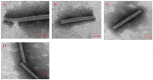
Methodology/principal findings: The crystal structure of a genetically engineered TMV CP was resolved at 3.06 Å. For the genetically engineered TMV CP, a six-histidine (His) tag was introduced at the N-terminus, and the C-terminal residues 155 to 158 were truncated (N-His-TMV CP(19)). Overall, N-His-TMV CP(19) protein self-assembled into the four-layer aggregate form. The conformations of residues Gln36, Thr59, Asp115 and Arg134 were carefully analyzed in the high radius and low radius regions of N-His-TMV CP(19), which were found to be significantly different from those observed previously for the helical and four-layer aggregate forms. In addition, the aggregation of the N-His-TMV CP(19) layers was found to primarily be mediated through direct hydrogen-bonding. Notably, this engineered protein also can package RNA effectively and assemble into an infectious virus particle.
Conclusion: The terminal sequence of amino acids influences the conformation and interactions of the four-layer aggregate. Direct protein-protein interactions are observed in the major overlap region when residues Gly155 to Thr158 at the C-terminus are truncated. This engineered TMV CP is reassembled by direct protein-protein interaction and maintains the normal function of the four-layer aggregate of TMV CP in the presence of RNA.
Conflict of interest statement
Figures




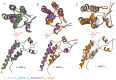

Similar articles
-
The development and application of new crystallization method for tobacco mosaic virus coat protein.Virol J. 2012 Nov 21;9:279. doi: 10.1186/1743-422X-9-279. Virol J. 2012. PMID: 23171808 Free PMC article.
-
Role of hexahistidine in directed nanoassemblies of tobacco mosaic virus coat protein.ACS Nano. 2011 Mar 22;5(3):1606-16. doi: 10.1021/nn1025719. Epub 2011 Feb 25. ACS Nano. 2011. PMID: 21361370
-
New Strategies and Methods to Study Interactions between Tobacco Mosaic Virus Coat Protein and Its Inhibitors.Int J Mol Sci. 2016 Feb 26;17(3):252. doi: 10.3390/ijms17030252. Int J Mol Sci. 2016. PMID: 26927077 Free PMC article.
-
Switching in the self-assembly of tobacco mosaic virus.Adv Biophys. 1990;26:157-85. doi: 10.1016/0065-227x(90)90011-h. Adv Biophys. 1990. PMID: 2082726 Review.
-
Molecular assembly of tobacco mosaic virus in vitro.Adv Biophys. 1986;22:95-149. doi: 10.1016/0065-227x(86)90004-3. Adv Biophys. 1986. PMID: 3551520 Review.
Cited by
-
Chemical and Biological Investigations of Antiviral Agents Against Plant Viruses Conducted in China in the 21st Century.Genes (Basel). 2024 Dec 23;15(12):1654. doi: 10.3390/genes15121654. Genes (Basel). 2024. PMID: 39766921 Free PMC article. Review.
-
Binding studies between cytosinpeptidemycin and the superfamily 1 helicase protein of tobacco mosaic virus.RSC Adv. 2018 May 23;8(34):18952-18958. doi: 10.1039/c8ra01466c. eCollection 2018 May 22. RSC Adv. 2018. PMID: 35539684 Free PMC article.
-
New chalcone derivatives: synthesis, antiviral activity and mechanism of action.RSC Adv. 2020 Jun 26;10(41):24483-24490. doi: 10.1039/d0ra03684f. eCollection 2020 Jun 24. RSC Adv. 2020. PMID: 35516226 Free PMC article.
-
Synthesis, Antiviral Bioactivity of Novel 4-Thioquinazoline Derivatives Containing Chalcone Moiety.Molecules. 2015 Jun 29;20(7):11861-74. doi: 10.3390/molecules200711861. Molecules. 2015. PMID: 26132908 Free PMC article.
-
Design, synthesis, antiviral bioactivities and interaction mechanisms of penta-1,4-diene-3-one oxime ether derivatives containing a quinazolin-4(3H)-one scaffold.BMC Chem. 2019 Mar 25;13(1):34. doi: 10.1186/s13065-019-0547-1. eCollection 2019 Dec. BMC Chem. 2019. PMID: 31384782 Free PMC article.
References
-
- Stubbs G, Kendall A (2012) Helical viruses. Adv Exp Med Biol 726: 631–58. - PubMed
-
- Bloomer AC, Butler PJG (1986) Tobacco mosaic virus structure and self-assembly: The plant viruses. In: Regenmortel MHV, Fraenkel-Conrat H. editors. New York: Plenum Press. 19–57.
-
- Mandelkow E, Holmes KC, Gallawitz U (1976) A new helical aggregate of tobacco mosaic virus protein. J Mol Biol 102: 265–285. - PubMed
-
- Jonathan P, Butler G, Durham AC (1977) Tobacco mosaic virus protein aggregation and the virus assembly. Adv Protein Chem 31: 188–251. - PubMed
-
- Mandelkow E, Stubbs G, Warren S (1981) Structures of the helical aggregates of tobacco mosaic virus protein. J Mol Biol 152: 375–386. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

